Abstract
P3DB (http://www.p3db.org/) provides a resource of protein phosphorylation data from multiple plants. The database was initially constructed with a dataset from oilseed rape, including 14 670 nonredundant phosphorylation sites from 6382 substrate proteins, representing the largest collection of plant phosphorylation data to date. Additional protein phosphorylation data are being deposited into this database from large-scale studies of Arabidopsis thaliana and soybean. Phosphorylation data from current literature are also being integrated into the P3DB. With a web-based user interface, the database is browsable, downloadable and searchable by protein accession number, description and sequence. A BLAST utility was integrated and a phosphopeptide BLAST browser was implemented to allow users to query the database for phosphopeptides similar to protein sequences of their interest. With the large-scale phosphorylation data and associated web-based tools, P3DB will be a valuable resource for both plant and nonplant biologists in the field of protein phosphorylation.
INTRODUCTION
Protein phosphorylation is the most studied posttranslational modification that controls the dynamic behaviors and decision processes in cells of various organisms. In recent years, large-scale studies on protein phosphorylation based on mass spectrometry have been conducted on different organisms. Most of these studies were undertaken in mammals and bacteria (1–5). Some of them were carried out in plants (6–8).
As a result, a number of phosphorylation databases emerged, most of which focus on mammalian and prokaryotic systems. Phospho.ELM (9) contains verified eukaryotic phosphorylation sites, but most are from mammals. PHOSIDA (10) contains large-scale phosphorylation data in Homo sapiens, Bacillus subtilis and Escherichia coli. PhosphoSitePlus (http://www.phosphosite.org/) contains curated phosphorylation sites mainly in vertebrates. Some of the phosphorylation databases focus on plants. PlantsP (11) contains phosphorylation data on a few different plants, but it focuses on the annotation of plant protein kinases and protein phosphatases. PhosphAt (12) provides a database of phosphorylation sites collected from current literature solely for the model organism Arabidopsis thaliana.
P3DB is unique in that it provides a resource of protein phosphorylation sites from various plant sources and contains multiple embedded search capacities for querying the database. By collecting and annotating plant phosphorylation data from different plant sources in a single database as a ‘one-stop’ shop, we anticipate P3DB that will serve as a useful resource not only for molecular biologists to study protein phosphorylation in plants and nonplant systems by comparison, but also for bioinformaticians to develop computational prediction tools on protein phosphorylation.
DATA COLLECTION
The database was constructed with a dataset from oilseed rape (Brassica napus var. Reston) developing seed obtained using a combination of data-dependent neutral loss and multistage activation on an LTQ linear ion trap liquid chromatography tandem mass spectrometry system. Details on the experimental design, which are available on the website (P3DB V1.0 release note), and the associated results and data analysis will be published elsewhere (Agrawal et al., unpublished results). The dataset includes 14 670 nonredundant phosphorylation sites (8350 phosphoserine sites, 4750 phosphothreonine sites and 1567 phosphotyrosine sites) from 6382 substrate proteins, representing the largest collection of plant phosphorylation data to date. Experimental details about each phosphopeptide, such as charge state, cross-correlation score, peptide probability, spectrum count, spectrum plot, etc., are available in the database.
More protein phosphorylation data are being deposited into this database from recently completed large-scale studies of A. thaliana (Columbia) and soybean (Glycine max var. Maverick). Phosphorylation data from other, previous investigations are also being integrated into the P3DB. For example, we have integrated a dataset published in Ref. (8) into the P3DB. Users are also encouraged to submit their own plant phosphorylation data to P3DB. Submitted data will be displayed according to the current database format with full credit given to the submitting investigators.
ACCESS TO THE DATA
Protein phosphorylation data are stored in a MySQL relational database. With a PHP-based web graphical interface, the phosphorylation data in the database are downloadable, browsable and searchable. The entire dataset can be downloaded in a tab-delimited format. A user can browse the annotated phosphoproteins by organisms or by gene ontology categories (13). A user can search for phosphoproteins by protein identifiers (NCBI GI numbers, UniProt accession numbers or RefSeq accession numbers) or protein descriptions, and search for phosphopeptides by peptide sequences. The main page of the search result lists all phosphoproteins/peptides meeting the searching criteria and gives some brief information, such as protein accession, protein description, source organism, consensus score, spectrum count, etc. The user can sort the result table according to different criteria, e.g. sort the phosphoproteins according to spectrum count from high to low. From the search result page, the user can navigate among pages of phosphoproteins, phosphopeptides and phosphorylation sites. The phosphoprotein page gives the details on the substrate protein, including the protein sequence with phosphorylation sites linked. Clicking on a phosphorylation site will display its detailed information, such as its surrounding amino acids (+/−10) and a list of phosphopeptides that contain this phosphorylation site. The information on each phosphopeptide is hidden by default to simplify entry page appearance. Clicking on ‘Show details’ presents the information about the peptide and clicking on ‘More’ takes the user to the phosphopeptide page which contains additional information about the peptide.
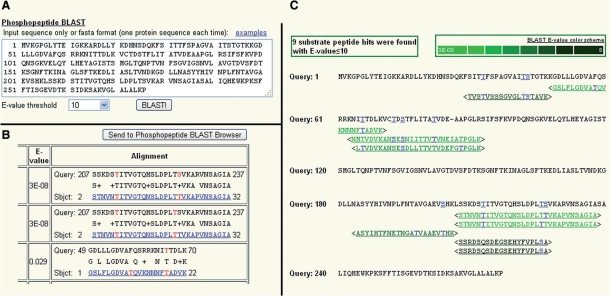
Another useful feature on the website is the phosphopeptide BLAST utility as shown in Figure 1. By uploading a protein sequence as in Figure 1A and querying it against the database using BLAST, a user can identify all the peptides in the query sequence that match one or more phosphopeptides in the database (according to a user-defined E-value cutoff). In the BLAST result page as Figure 1B, the BLAST alignments are displayed with links to the phosphopeptides and phosphorylation sites. In addition, to graphically representing phosphopeptide BLAST results, we developed a tool to view phosphopeptide BLAST results. Figure 1C shows one example after submitting a phosphopeptide BLAST result to this tool by clicking on ‘Send to Phosphopeptide BLAST browser’ in Figure 1B. All the BLAST alignments are displayed with an E-value color scheme so that the user can know how similar the peptides in the query sequence are to the phosphopeptides. In addition, each residue in the query sequence that is aligned to one or more phosphorylation sites in the matching phosphopeptides is explicitly colored and hyperlinked, if it is serine, threonine or tyrosine. A user can also submit a protein query sequence directly to this tool under the ‘Tools’ menu. The BLAST utility and BLAST result browser does not aim to explicitly predict phosphorylation sites or phosphorylation motifs in the query protein sequences, but does help the user to gain some related biological meaning about the query sequences. For example, if a user has a human phosphoprotein in hand and is interested to know whether similar phosphopeptides exist in plants, he/she may find this tool useful. Alternatively, if a user wants to know whether a plant protein contains phosphorylation sites, this tool may help him/her to gain some knowledge of the empirical evidence for phosphorylation based on the related sequences from the database in a conservative or semi-conservative manner.
Figure 1.
Phosphopeptide BLAST example (Query sequence: Arabidopsis ATP binding protein; Dataset: Oilseed rape phosphopeptides). (A) User interface for inputting a query protein sequence and selecting E-value threshold. (B) BLAST result page (partial). The BLAST alignments with their E-values are displayed. The matching phosphopeptides are hyperlinked to the corresponding phosphopeptide pages which show their detailed information. The phosphorylation sites in the matching peptides are also hyperlinked and colored as red. (C) BLAST result browser. Sequences following ‘Query: #’ are from the query protein sequence. Sequences between angle brackets (<>) are the matching phosphopeptides. They are hyperlinked to the phosphopeptide pages. The peptide sequences are rendered with different colors to show the different E-values of hits as indicated in the BLAST E-value color scheme legend. Each phosphorylation site in each phosphopeptide is also hyperlinked and colored as blue, and so is the corresponding residue in the query sequence if it is ‘S’(serine), ‘T’(threonine) or ‘Y’(tyrosine).
FUTURE DIRECTION
Deposit phosphorylation datasets from large-scale studies of A. thaliana and soybean, which are in the process of being annotated.
Integrate more plant phosphorylation datasets from other investigators into P3DB and continue updating the database with new advances in mining and prediction analysis of plant phosphorylation.
Integrate information on phosphorylation motifs and protein kinase specificity.
Integrate additional information on protein phosphorylation data, such as Pfam domain, cross-species conservation data, pathway information, etc.
Improve the current utilities and implement more tools, such as advanced search tool for querying by a user-defined combination of different criteria.
Predict protein structures of phosphoproteins and highlight phosphorylation sites in a web-based protein structure viewer.
FUNDING
National Science Foundation (grant number DBI-0604439 to J.T.). Funding for open access charges: National Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. Due to space constraints the authors regret they could not cite all relevant research articles.
REFERENCES
- 1.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 2.Villén J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl Acad. Sci. USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl Acad. Sci. USA. 2007;l 104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl Acad. Sci. USA. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal GK, Thelen JJ. Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol. Cell Proteomics. 2006;5:2044–2059. doi: 10.1074/mcp.M600084-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 2008;4:193. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites–update 2008. Nucleic Acids Res. 2008;36(Database issue):D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tchieu JH, Fana F, Fink JL, Harper J, Nair TM, Niedner RH, Smith DW, Steube K, Tam TM, Veretnik S, et al. The PlantsP and PlantsT functional genomics databases. Nucleic Acids Res. 2003;31:342–344. doi: 10.1093/nar/gkg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008;36(Database issue):D1015–D1021. doi: 10.1093/nar/gkm812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32(Database issue):D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]