Abstract
The Proteolysis MAP (PMAP, http://www.proteolysis.org) is a user-friendly website intended to aid the scientific community in reasoning about proteolytic networks and pathways. PMAP is comprised of five databases, linked together in one environment. The foundation databases, ProteaseDB and SubstrateDB, are driven by an automated annotation pipeline that generates dynamic ‘Molecule Pages’, rich in molecular information. PMAP also contains two community annotated databases focused on function; CutDB has information on more than 5000 proteolytic events, and ProfileDB is dedicated to information of the substrate recognition specificity of proteases. Together, the content within these four databases will ultimately feed PathwayDB, which will be comprised of known pathways whose function can be dynamically modeled in a rule-based manner, and hypothetical pathways suggested by semi-automated culling of the literature. A Protease Toolkit is also available for the analysis of proteases and proteolysis. Here, we describe how the databases of PMAP can be used to foster understanding of proteolytic pathways, and equally as significant, to reason about proteolysis.
INTRODUCTION
Regulatory proteolysis is an important and unique type of posttranslational modification because it is irreversible. Proteolysis is essential to almost all fundamental cellular processes including proliferation, death and migration (1–4). Equally as important, mis-regulated proteolysis can cause diseases ranging from emphysema (5) and thrombosis (6), to arthritis (7) and Alzheimer's (8). There are a number of online resources containing information on proteases including SwissProt (the oldest), HPRD (human protein reference database) (9) and UniProt (10). The best known protease resource, MEROPS (11) provides a ‘gold standard’ in protease classification, and basic information on almost 100 000 proteases. However, none of these resources address the predictive modeling of proteolytic events or the analysis of proteolytic networks. The Proteolysis MAP (PMAP) reasoning environment was established with these objectives in mind. It aims to take advantage of information available in public databases, results from experiments, the users own imagination to perform specific queries, and then brings these elements together to efficiently address dependent queries like:
I know of a protease that is up-regulated in a specific disease. What are the likely substrates for this protease that could drive the pathology?
I have identified a protein that is cleaved during a biological event, and I know the position of the cut site. Which proteases are likely to be responsible for this cleavage?
I know of a protease and its substrate that are necessary for a biological event. What other proteins might associate with these two in a regulatory network?
I have identified a cut site within a protein, but I have indications that such cleavage is regulated. Are there other posttranslational modifications within the protein that are likely to regulate proteolysis?
I have found two compounds that interfere with a proteolytic pathway in apparently different time-regimes. Can I quantitatively model the pathway to identify their potential protein targets and gain insight into differences in their behavior?
I have a virus protease that participates in maturation of viral proteins and want to know if it may also have some human substrates.
Within the context of PMAP, these queries are addressed by bringing together seemingly unrelated or physically disconnected information that is now stored in a set of five interacting databases: ProteaseDB, SubstrateDB, CutDB, ProfileDB and PathwayDB. Their integration is sufficient to make them of great value to the research community.
DATABASES OF PMAP
ProteaseDB
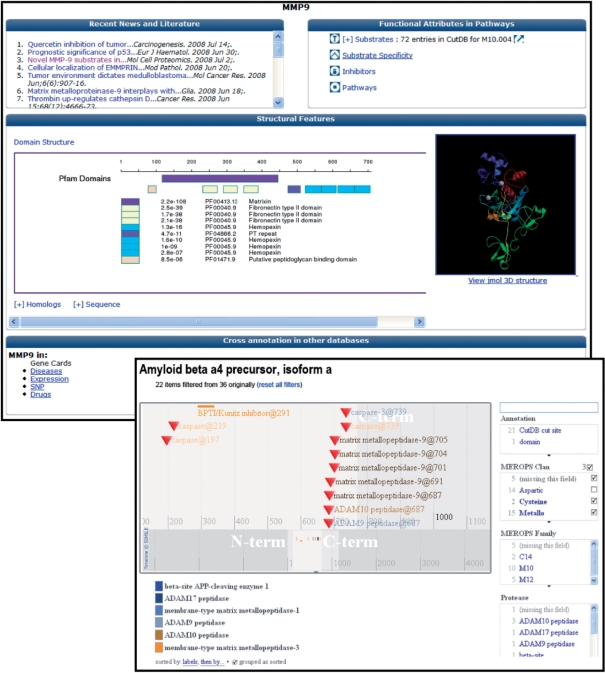
ProteaseDB contains basic information for a set of ∼150 human proteases deemed to be of high interest. The database actually contains more than 45 000 proteases acquired from MEROPS, and information on this comprehensive set of proteases will be displayed in the form of Molecule Pages as the site matures. The information is stored in a MySQL database and presented as a Molecule Page on a web server (Figure 1, top, as an example). Each Molecule Page displays a comprehensive set of annotations on 15 different features of proteases, all acquired from an automated protein annotation pipeline run on a Linux cluster. These features include predictions of PFAM domain structure, secondary structure, transmembrane regions, signal peptides and disordered regions. Among the methods used in the protein annotation pipeline are: BLASTP, protein sequence homology search (12); Hmmer, hidden Markov model search (13); TMHMM, transmembrane helices prediction (14); SignalP, signal peptide prediction (15); Jnet, secondary structure prediction (16); Coils, coiled-coil region prediction (17); Seg, low complexity segment identification (18); MODELLER, homology modeling of protein 3D structures (19). The 3D structure, or high-resolution model, of each protease is presented within a Jmol viewer capable of querying and displaying many structural features. Each Molecule Page is also linked to external sites (MEROPS, PDB, PubMed, GeneCards and the GNF SymAtlas) from which data was collected and where additional information can be found. Molecule Pages present ‘Recent News and Literature’, which shows selected articles from PubMed related to the protease via a dynamic web services query (see Supplementary Material). If entries for a protease are present in CutDB, a simplified list of substrates is present, thus connecting ProteaseDB to both CutDB and SubstrateDB.
Figure 1.
Protease Molecule Page (top) and Substrate Molecule Page (bottom). The Protease Molecule Page shows recent news and literature of the protease (top left), known proteolytic events (top right), domain location and structure view (middle panel), as well as a cross annotation in other databases section (bottom). A graphics interface of SubstrateDB (bottom) is shown here displaying domains and experimental protease cut sites for the human amyloid β A4 protein. A total of 35 protease cut sites are filtered via selection boxes on the right. The display makes co-location of many features visible. The annotation displayed is linked back to sources (CutDB, ProteaseDB, UniProt, etc.) for more detailed annotation. The graphics display is synchronized with a text table of the same data, which can also be downloaded.
SubstrateDB
SubstrateDB contains molecular information on documented protease substrates in CutDB. SubstrateDB differs from ProteaseDB in that it is designed to map a variety of annotations onto the primary sequence of known or predicted substrates (Figure 1, bottom). The database architecture allows other posttranslational modifications to be mapped onto the primary sequence of the substrate. Documented protease cleavage sites are visible within the context along with all other potential regulatory modifications.
To achieve this type of dynamic mapping, and to keep it current, SubstrateDB dynamically collects data from PMAP, UniProt and InterPro [via web services APIs (20)]. Substrate pages are constructed at the request of the user. To reduce the wait-time for gathering and integrating information, the system retains a dynamic cache of data from prior requests. Information within SubstrateDB is displayed onto the primary structure of the substrate using a Java-script driven interface adapted from the Simile project hosted by CSAIL (MIT/Broad Institute, (http://simile.mit.edu/timeline/). We have adapted the Simile's Timeline interface to plot data along a one-dimensional axis corresponding to the primary sequence of the substrate. The Timeline interface allows users to dynamically select subsets of annotation for display.
CutDB
CutDB is a database of individual proteolytic events (cleavage sites) culled from the literature. It represents one of the first systematic efforts to build a collection of documented proteolytic events (21), and as such, is the largest database of proteolytic events in the world. The database has more than 5300 annotated proteolytic events that occur within 1702 protein substrates. The annotated proteolytic events are enacted by 180 serine, 164 metallo, 108 cysteine and 61 aspartic proteases. The seed data for CutDB (∼2000 substrates) was extracted from MEROPS, HPRD and UniProt. Then ∼3000 additional substrates were extracted from original articles by a combination of directed literature searches and readings by human experts.
Each proteolytic event is annotated as a cut site location within the primary sequence of the protein substrate. The residues spanning the cut site are highlighted. Events are annotated with core information including the molecular identity of the protease and substrate which are each linked to their pages within ProteaseDB and SubstrateDB. Each event is also connected to Literature Track, which has information of the articles (PubMed IDs including links) used to annotate the event. When available, other features associated with the event are also stored in CutDB. These include information on the method by which the event was detected, the potential consequences of the event, relevant cofactors, associated pathways, cell compartments where the event takes place, cell lines where the event is observed and information linking the event to any process or disease.
Any registered user can curate the content of CutDB by adding new events, fixing errors or adding comments. The comment section is divided into several categories, such as ‘discussion’, ‘hypothesis’, ‘drugs in development’ and ‘other comment’. Manual annotation is already being conducted using the web interface by users throughout the world. This database will soon be extended to include proteolytic events detected by mass spectrometry and by statistical predictions (see ProfileDB section).
ProfileDB
ProfileDB is a protease specificity profiling database that contains information on substrate recognition from phage display experiments and other systematic studies, like peptide libraries. It has tools for using these data to predict protease cleavage sites in proteins and a screening engine for searching potential substrates in the whole human proteome. This database also supports community-based input and analysis. ProfileDB allows the user to search the database for a particular data set and submit specificity profiling data, so that the results from this work become easily accessible to other scientists. Using the database interface, it is easy to create protease specificity models (position weight matrix or PWM) and predict substrate cleavage sites using other specificity models or specificity motifs in a single substrate or fasta file. The methods we use to compute PWM and to predict protease cleavage sites have been previously described (22). An important feature of our proteome-wide search is the ability to filter proteome prediction by multiple criteria, like protein localization, cleavage site secondary structure or the presence of disordered regions.
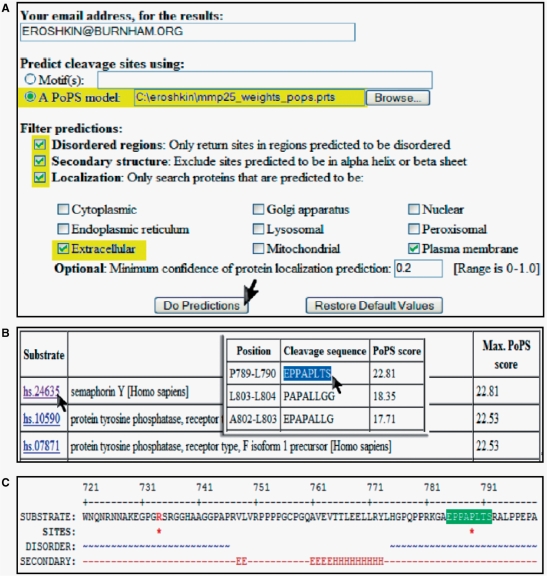
A simple scenario of how a user submits a proteome-wide search for substrates is shown in Figure 2A. After selection of the ‘do predictions’, the user will receive the result table of prioritized (by PWM score) potential substrates (Figure 2B) and, finally, the details of the cleavage site, including the amino acid sequence, protein secondary structure, disorder regions and Pfam domains (Figure 2C). The front page of the database presents raw profiling data for seven matrix metalloproteinases (MMPs, studied by The Center on Proteolytic Pathways), prebuilt PWM matrices for each of MMPs and proteome-wide prediction results.
Figure 2.
ProfileDB proteome-wide substrate search. (A) Initial page where the user selects options for proteome screening. (B) A results table generated from the proteome-wide screening with a detailed output for a particular protein. (C) Presentation of the cleavage site on the amino acid sequence with protein sequence features (secondary structure and disorder regions location).
PathwayDB
PathwayDB is the most recent addition to PMAP and is still in its formative stages. This database contains three types of information on proteolytic pathways and networks: (i) pathways reconstructed in silico using proteolytic event(s) as an initial seed, (ii) protease inferred networks reconstructed by a focused lexical application of Cytoscape (23) and (iii) scenarios of the function of proteolytic pathways derived from rule-based modeling. PathwayDB stores and displays, upon user request, pathways previously constructed from each of these scenarios. PathwayDB is a community-driven database where a registered user can create or modify pathways. Though the current content of PathwayDB is limited, it is continuously being expanded by the group of curators at The Center on Proteolytic Pathways, and by outside users.
Event-driven reconstruction
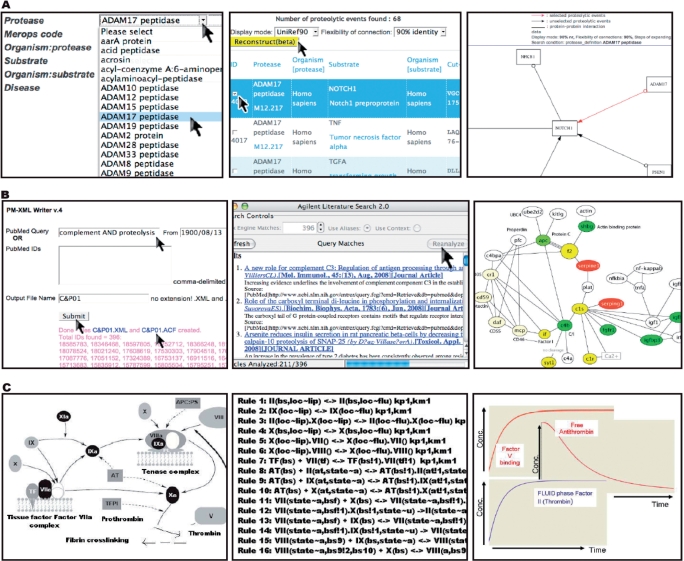
Single-order proteolytic pathways are reconstructed in silico by using proteolytic events from CutDB as a seed. The user can select an event, or a series of proteolytic events, and the system constructs a network diagram in which the nodes correspond to proteases and substrates (Figure 3A). Hence, one can easily visualize all of the substrates for a given protease, or even all of the proteases that cleave a given substrate. These networks can be expanded to another level by including protein–protein interaction data taken from the IntAct database and homology information from UniRef (24). This allows a vastly simplified visualization of the network and eliminates a great deal of redundancy. In the network diagrams that are generated, the substrate names are converted into HUGO gene symbols (25), which are stored in NCBI RefSeq records. Reconstructed network can be submitted for storage to PathwayDB (with appropriate title and description given by the author) and retrieved by any user.
Figure 3.
Three types of information in PathwayDB. (A) Automated network reconstruction for one proteolytic events caused by ADAM17 peptidase and Notch1 preproprotein. Left panel: from CutDB front page, users search for the events. Middle panel: selection of events to reconstruct networks. Users can also define three parameters to show the network diagrams: (i) ID display mode; (ii) flexibility of connection, the nodes are connected within given threshold of sequence similarity; and (iii) steps of expanding, the edges are extended by given extension level. Right panel: the reconstructed pathway. The red arrows indicate the selected events. The black arrows indicate expanded events. The nonarrows edges are protein–protein interactions. (B) Proteolytic inferred networks. Left panel: PubMed-XML writer, a web service for extracting Medline abstracts, is used to delimit lexical queries, for example ‘complement AND proteolysis’. These abstracts are then assembled in XML format and submitted to Agilent Literature Search, acting as plug-in to Cytoscape (middle panel), which parses sentences containing ontologically known entities (such as gene and protein names). Cytoscape takes these gene/protein names and builds interactive networks as hyper-graphs. Right panel: hyper-graph generated for one ‘Network of the month’ entitled ‘Complement activation and regulation 01’. This network captured five proteases (nodes in yellow) that interplay with six substrates (nodes in green), four cofactors (nodes in light yellow), two inhibitors (nodes in red) and other binding proteins such as C4BP (white nodes). (C) Rule-based modeling of network function. Left panel: schematic representation of major events involved in coagulation. Middle panel: rules for coagulation cascade. Right panel: modeling of production/consumption profiles of individual components of the pathway using an in-house developed rule-based model for coagulation. In the plot, the x-axis denoted the time of simulations (100 iterations) and the y-axis is the molecular concentrations in nanomolar (0–100 000).
Proteolytic inferred networks
PathwayDB contains inferred networks in the form of a ‘Network of the month’. These networks are developed using a focused application of Cytoscape (23). Ultimately, PMAP will provide users with the opportunity to construct networks in the same manner. The ‘Network of the month’ is a natural language processing (NLP)-based and author-driven network of molecular events including proteolysis of interest. It is extracted from natural language text such as PubMed abstracts, and is represented in a hierarchical hyper-graph data structure (23,26). By adding this new capability to Cytoscape, we are able to extract information on proteolytic events from Medline and automatically reconstruct protease inferred networks. The example network shown in Figure 3B was constructed starting from a simple two-word query (e.g. ‘complement AND proteolysis’) to PubMed. GO names found in the resulting 396 MEDLINE abstracts were imported into Cytoscape to generate the literature-supported inferred network.
Rule-based modeling of network function
In some cases, a great deal of knowledge already exists for a network or pathway, and the user is interested in knowing the outcome of perturbations to the system. PathwayDB has implemented a rule-based approach using the BioNetGen framework (27) to evaluate such queries (Figure 3C). Initially, all distinct states of reactant species and complexes are calculated using the BioNetGen framework. Then, a quantitative model is generated that encompasses possible reactions implied by the nature of interactions, using the initial reactant species and an expanded set of molecular species (complexes). Individual reactions in the previous step are further parameterized with rate constant data coming from the corresponding rules. The user has the capability of viewing the detail parameters of the underlying default model. In addition, the rules and parameters of the default model can be modified to generate advanced models. We developed an upload system, where the user can submit a rule-based model with description, data files and a model diagram to PathwayDB. The uploaded model immediately becomes available for other users of the system. In the future, we aim to generate a framework for using these pathway models to assist the user in making functional hypotheses in specific proteolytic pathways.
APPLICATIONS
There are numerous applications of PMAP. Subsequently, we briefly outline how the databases of PMAP can be applied to address point-by-point the dependent queries put forth in the Introduction section:
Query (i)
Workflow
First the user can search CutDB, to determine if there are any substrates for the protease that have been reported in the literature. In the absence of any known substrates, the user can use ProfileDB to determine if the substrate recognition profile of the active site has been determined. If so, ProfileDB can be used to identify likely, or predicted, substrates for the protease of interest.
Query (ii)
Workflow
The user can perform a query of CutDB to determine if the protein is a known substrate for any proteases. If there are multiple proteases that cut the protein, then the information in CutDB will point to the protease responsible for a particular cut site. If there is no documented protease that cleaves the protein of interest, then the user can search ProfileDB to determine if the protein is a predicted substrate for any protease.
Query (iii)
Workflow
The user can take two approaches toward this question. First, the user can search CutDB for all the substrates for the protease, and all of the potential proteases that cleave the substrate. The user can ask PathwayDB to link all of these proteases and substrates in a network map. The user can also ask CutDB to extend the association map to include known protein–protein interactions. Alternatively, the user can request that the PMAP team apply the inferred network feature of PathwayDB to identify abstracts within PubMed that link the protease and substrate or any other useful search term. Then, PathwayDB can be directed to funnel this information into Cytoscape to arrive at a network map. We anticipate that the number of these inferred pathways stored within the database will grow as more users take advantage of the site, but consideration is also being given to providing a semi-automated workflow of this type to the user.
Query (iv)
Workflow
To address this question, the user can access SubstrateDB and pull forward the page associated with the substrate of interest. Here, the user can paint the substrate with a wide range of detailed molecular information, including the position of domains and posttranslational modifications.
Query (v)
Workflow
The user can apply the rule-based modeling feature of PathwayDB, and apply various constraints, including those that occur in response to the inhibitors. Rule-based modeling within PathwayDB will allow the user to generate perturbed models. These models then can be further studied using for example virtual cell environment (28) to decipher quantitative changes of the underlying pathway and thus allowing the user to ‘visualize’ the potential consequences of perturbation.
Query (vi)
Workflow
The user can start with the submission of known virus cleavage sites to ProfileDB, followed by development of a protease specificity model using known cut sites and prediction of potential substrates in an entire human proteome. The user can then look for tissue-related, physiologically relevant proteins in a prioritized list and check experimentally if they can be cleaved in vitro.
PMAP ARCHITECTURE AND IMPLEMENTATION
The ProteaseDB and SubstrateDB applications use the Catalyst web framework, whereas CutDB uses the Ruby on Rails web application framework. Other tools use a variety of frameworks and languages. The PMAP application that integrates components and links these components together was build using the Perl-based Catalyst web framework. Catalyst web framework allows us to separate the components of our system into three parts: the Model, the View, and the Controller (MVC). The Model represents the data of the application, the View specifies the user interface, and the Controller handles communication among all elements of the application.
Simple Object Access Protocol (SOAP)-based web services have been used to integrate the different database applications in PMAP. SOAP has been effective in allowing simple, reliable connections between diverse resources. In one case, to list substrates for a particular protease (from Ruby on Rails), and to grab the most current literature from PubMed related to that protease (from Windows ASP) and deliver these integrated results within ProteaseDB page (via Perl, Catalyst).
In CutDB and PathwayDB, all frameworks for the web interface are implemented using Ruby on Rails. The database in the background is MySQL. The web server is Lighttpd. The network diagram is generated by Graphviz.
In the future, we will continue to programmatically integrate the separate databases of the PMAP project, add user annotation, and develop it in such a way that we will be able to add functionality and maintain code as efficiently as possible.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
National Institutes of Health (RR020843, CA108959, CA30199). Funding for open access charge: National Institutes of Health (RR020843, CA108959, CA30199).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank many at the Burnham Institute for Medical Research for the data curation and Dr Weizhong Li for providing protein annotation pipeline. We thank Dr Christina Niemeyer for her editorial assistance and critical review of the manuscript.
REFERENCES
- 1.King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 2.Kudo NR, Wassmann K, Anger M, Schuh M, Wirth KG, Xu H, Helmhart W, Kudo H, McKay M, Maro B, et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–146. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 4.Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science. 2004;306:108–111. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]
- 5.Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur. Respir. J. 2003;22:672–688. doi: 10.1183/09031936.03.00040703. [DOI] [PubMed] [Google Scholar]
- 6.Carrell RW, Owen MC. Plakalbumin, alpha 1-antitrypsin, antithrombin and the mechanism of inflammatory thrombosis. Nature. 1985;317:730–732. doi: 10.1038/317730a0. [DOI] [PubMed] [Google Scholar]
- 7.Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 8.Haass C, De Strooper B. The presenilins in Alzheimer's disease—proteolysis holds the key. Science. 1999;286:916–919. doi: 10.1126/science.286.5441.916. [DOI] [PubMed] [Google Scholar]
- 9.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawlings ND, Tolle DP, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 14.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 15.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 16.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 17.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 18.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 19.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 20.Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi Y, Eroshkin A, Gramatikova S, Gramatikoff K, Zhang Y, Smith JW, Osterman AL, Godzik A. CutDB: a proteolytic event database. Nucleic Acids Res. 2007;35:D546–D549. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd SE, Pike RN, Rudy GB, Whisstock JC, Garcia de la Banda M. PoPS: a computational tool for modeling and predicting protease specificity. J. Bioinform. Comput. Biol. 2005;3:551–585. doi: 10.1142/s021972000500117x. [DOI] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 25.Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res. 2006;34:D319–D321. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vailaya A, Bluvas P, Kincaid R, Kuchinsky A, Creech M, Adler A. An architecture for biological information extraction and representation. Bioinformatics. 2005;21:430–438. doi: 10.1093/bioinformatics/bti187. [DOI] [PubMed] [Google Scholar]
- 27.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. BioNetGen: software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics. 2004;20:3289–3291. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 28.Slepchenko BM, Schaff JC, Macara I, Loew LM. Quantitative cell biology with the virtual cell. Trends Cell Biol. 2003;13:570–576. doi: 10.1016/j.tcb.2003.09.002. [DOI] [PubMed] [Google Scholar]