Abstract
SpBase is a system of databases focused on the genomic information from sea urchins and related echinoderms. It is exposed to the public through a web site served with open source software (http://spbase.org/). The enterprise was undertaken to provide an easily used collection of information to directly support experimental work on these useful research models in cell and developmental biology. The information served from the databases emerges from the draft genomic sequence of the purple sea urchin, Strongylocentrotus purpuratus and includes sequence data and genomic resource descriptions for other members of the echinoderm clade which in total span 540 million years of evolutionary time. This version of the system contains two assemblies of the purple sea urchin genome, associated expressed sequences, gene annotations and accessory resources. Search mechanisms for the sequences and the gene annotations are provided. Because the system is maintained along with the Sea Urchin Genome resource, a database of sequenced clones is also provided.
INTRODUCTION
The enterprise which resulted in the sequencing of the purple sea urchin genome began with support from the sea urchin research community and the Stowers Institute for Medical Research. The early efforts were directed to the production of cDNA and genomic libraries that would benefit laboratory studies of this widely used biomedical research model. These early library resources were used as the subject of a variety of medium-throughput studies. Details of the sequencing efforts and availability of resources were presented on a web site: Sea Urchin Genome Project (SUGP; http://sugp.caltech.edu/SUGP). In addition to cDNA studies (see below), an increasing number of bacterial artificial chromosome (BAC) clones were sequenced and made available at this site. Eventually these resources became the basis for the Sea Urchin Genome Sequencing Project (http://www.genome.gov/11008265). The Baylor College of Medicine Human Genome Sequencing Center took the lead in the genome sequencing project (http://www.hgsc.bcm.tmc.edu/projects/seaurchin/).
The same rationale for sequencing the sea urchin genome, the utility as a research model, holds for the establishment of a thorough information system for this species. Gene discovery and characterization is much more efficient with a genome sequence in hand and high-throughput approaches soon emerge from the complete sequence available for a genome. Since the presentation of the genome in 2006 (1), the number of gene annotations has steadily increased. There are well over 10 000 annotations completed and listed in SpBase. Although the amount of expression information is as yet incomplete, these data have aided in gene discovery, too [see Ref. (2) for example].
The sea urchin genome sequence presents some unique problems for the bio-informatician. The high degree of polymorphism in the genome required special considerations in the assembly process (3). The draft sequence is a mosaic of two haplotypes estimated from the assembly to differ by about 2%. Because no physical or genetic map is available for the purple sea urchin, the highest level of organization is the individual scaffold. Genome database software is not optimized to handle ∼29 000 units instead of the 5–50 chromosomal units presented for genomes with mapping information. The first manifestation of this problem is slow responses in genome browsers and search functions.
THE DATA
Species
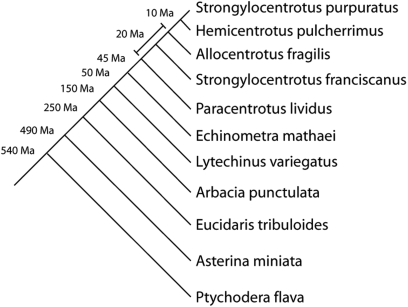
Within the echinoderm clade, regular echinoids (sea urchins) have been extensively used as biomedical research models due to their ease of handling and availability. Furthermore they are an excellent group of species for comparative genomics (Figure 1). Their fossil record is well-characterized back to the pre-Cambrian and molecular phylogenetic techniques have been extensively explored in the group (4–7). In addition to the reference species, Strongylocentrotus purpuratus (purple sea urchin), the database includes sequence information from other echinoderms, including three species of sea urchins (S. franciscanus, Allocentrotus fragilis and Lytechinus variegatus). A lesser amount of sequence data is also available from two other sea urchins (Arbacia punctulata and Eucidaris tribuloides) as well as a sea star (Asterina miniata) and a hemichordate (Ptychodera flava). We expect to see additional sequence incorporated from these species as ongoing sequencing projects are extended.
Figure 1.
The phylogeny of species from the echinoderm clade discussed in the text. The divergence time of the branches are indicated to the left of the line. All except the most divergent species are regular sea urchins and A. miniata is a sea star and P. flava belongs to Hemichordata, the sister phylum to the echinoderms.
Genome Assembly Version_0.5
The first version of the purple sea urchin genome was deposited in GenBank in August of 2005 (3). It was assembled from about 7 million whole genome shotgun reads derived from plasmids with 2- to 6-kb inserts. The DNA came from a single male sea urchin that also provided the material for a large BAC library (8). The reads represent about 6× coverage of the genome which is estimated to be 800 Mb in size. They were produced and assembled at the Baylor College of Medicine, Human Genome Sequencing Center (http://www.hgsc.bcm.tmc.edu/projects/seaurchin/) using the Atlas assembler (9). To aid in the assembly, about 109 000 BAC ends were sequenced from two different libraries: one with inserts averaging 130–160 kb and one with inserts of 30–50 kb. The N50 of the contigs >1 kb is 10.18 kb and the N50 of the scaffolds is 47.98 kb. The N50 size is the length such that 50% of the assembled genome lies in blocks of the N50 size or longer. Due to the preliminary nature of the scaffolding, the size of this initial assembly is 180 Mb larger than the estimated genome size and it is estimated to have about 15% redundancy when compared to 25 high-quality BAC sequences (3). Still, the size of the contigs and scaffolds was sufficient for gene predictions since the size of an average sea urchin gene is about 10 kb. Because this assembly was used to prepare the official gene set (OGS), it is preserved in SpBase. It is available as a BLAST database for searching and displayed in a genome browser with other genomic features mapped to it (see below).
Genome Assembly Version2.1
In order to improve the assembly, additional sequencing of BAC inserts was undertaken (1,3). For a genome like the sea urchin one which is highly polymorphic, BAC clone sequencing offers an added advantage since each BAC insert is derived from a single haplotype. This strategy of combining 6× WGS with low-coverage (2×) BAC sequencing was proved successful for the Rat Genome Sequencing Project (10). For the sea urchin, an additional efficiency was obtained by using a matrix pooling technique to reduce the number of sequencing libraries needed (11). The BACs to be sequenced were derived from a minimum tiling path revealed by a restriction enzyme fingerprinting protocol. The Atlas assembler software was specifically developed to combine WGS and BAC reads. Each set of BAC reads is used to ‘fish’ for overlapping WGS reads and a local assembly is performed. The product is called an ‘eBAC’ and the eBACs are stitched together by overlap and mate pair bridging. The BAC reads from the other haplotype can be added to the assembly later in the process (3). Thus the result is a mosaic of two haplotypes. This assembly was submitted to GenBank on 18 October 2006 as Spur_v2.1. This assembly is available at SpBase as a searchable BLAST database and in a genome browser. An alternate assembly was produced at National Center for Biotechnology Information (NCBI) which used mRNA, EST, protein and paired read alignments to order and orient the Spur_v2.1 scaffolds.
Expressed sequence tags
Expressed sequence tags (ESTs) and well-studied cDNAs from the purple sea urchin have been accumulating since the advent of arrayed libraries. Gene discovery and genomic sequence analysis both profit from these high-throughput approaches. A number of EST studies have contributed to the large data set of expressed sequences that number in excess of 140 000 sequences. Several independent sequencing or clustering projects contributed to this collection (8,12; http://genome.wustl.edu; http://www.hgsc.bcm.tmc.edu/projects/seaurchin/; http://www.molgen.mpg.de/~ag_seaurchin/). Most of these concentrated on embryonic stages of development although the Sea Urchin Genome Sequencing Project at Baylor did sample a variety of life stages and cell type libraries. It is interesting to note that a total of 93 636 transcribed sequences yielded 15 291 clusters in the June 2006 purple sea urchin Unigene build #10 at NCBI. Of these only 577 contain mRNAs and the rest are compiled from ESTs. This could represents as much as three-fourth of the genes in the genome. The largest gap in the coverage of transcribed sequences lies in the larval and adult stages, the embryonic cDNAs are well sampled by the EST projects that have been mounted to date.
The gene models
The contigs and scaffolds of the Spur_0.5 assembly were judged of sufficient length to support the operation of gene prediction pipelines with a high degree of accuracy. Four different gene prediction algorithms were independently employed by individual groups to arrive at sets of gene models for eventual annotation: Gnomon at NCBI (13); FgenesH from Softberry (14,15); a Genescan approach [http://urchin.nidcr.nih.gov/blast/index.html; (16)] and the Ensembl gene prediction pipeline (17) run by BCM-HGSC at Houston. Consensus gene models from these sets of predictions were generated by a statistical approach embodied in a program called GLEAN (18). The final OGS included 28 944 models (1,3). A careful estimate comparing the annotated genes and the statistics from various expressed sequence collections yielded a final estimate of 23 300 genes in the purple sea urchin genome (1).
BAC sequences
Over the period of time that the SUGP was underway, many individual BAC clones identified by the gene sequences they contained were sequenced to an ordered and oriented draft sequence (8). The main purpose of this effort was to provide noncoding sequence near transcription units for comparative genomic analysis to find cis-regulatory modules. These sequences have been submitted to an automated scan that maps cDNA and other features onto the BAC sequences. These are displayed on a separate series of web pages organized by the gene of interest contained in the sequence.
Annotations
The automated analysis of the original 28 944 gene models predicted from the Spur_0.5 assembly was frozen for analysis and publication on 28 March 2006. A consortium of over 240 investigators and students volunteered to contribute manual annotations of these predictions into a SQL databse at BCM-HGSC (http://www.genboree.org/). Over 10 000 manual gene annotations have been submitted at the time of this writing. In turn they have been organized into a searchable annotation database and presented on the SpBase web site. In addition to a series of sequence coordinates on the assembly scaffolds, the predicted gene models are identified by similarity to ESTs, cDNAs or independently derived and translated protein sequence. Representative homologous matches where available from deuterostome species were employed to characterize gene models or in their absence protostome sequences. Missing gene features have been recalculated and added as well.
In a move unusual for a primary annotation effort, the annotation consortium chose to incorporate wherever available expression data for individual genes into the data set. This action was undertaken on the basis that data accumulation on the scale proposed by the consortium may not occur again for a long time. A formal list of embryonic, larval and adult structures as well as developmental times and stages are included in the input form for annotation data. In the process of accumulating these expression data, a whole-genome tiling array was hybridized to RNAs pooled from embryonic stages up to 48 h of development (19). The resulting analysis showed that approximately 11 000–12 000 genes are utilized in the embryo, consistent with previous estimates from mRNA excess hybridizations performed 25–30 years ago. The tiling array data were used to correct and authenticate several thousand gene models during the genome annotation process.
The expression data accumulated during the annotation process is particularly interesting and is a major part of the information we wish to preserve and present. For example, these data show that expression of about 52% of the entire protein-coding genes in the sea urchin genome occurs in the first 48 h of development, up to the mid-late gastrula stage, while 80% of the sea urchin regulome of transcription factor genes (other than zinc-finger genes) are likewise expressed by 48 h of embryogenesis (20).
THE DATABASES AND WEB SITE
In an effort to minimize software development, the SpBase information system was constructed using previously engineered open source software components. The components were designed and are maintained by the Generic Model Organism Database Project, or GMOD, a consortium supported by experienced genomic database enterprises including WormBase, FlyBase, MGI, SGD, Gramene, Rat Genome Database, EcoCyc and TAIR (http://gmod/org). The components developed there emerged from a formal list of general recommendations drafted by the genomics community at large and components continue to be released.
The assembled genome sequences and the genomic features mapped to the two genome assemblies are organized into a PostgreSQL database structured with a schema named Chado (21). This schema was first designed for the Drosophila genome. It has more recently been generalized and included in the GMOD software suite. That the schema reached it's stated goals to be generic and extensible are evident in the frequency with which it has been adopted (21,22). The distinctive features of the schema include the use of controlled vocabularies and a modular structure organized around biological functions. Although the sea urchin system does not presently contain any information on genetic strains and microarray data, the portion of the schema for genomic features is well used. We have established independent databases for the two assemblies in order to preserve the sequence information used to predict the gene models and the most recent assembly, respectively. Because the Chado schema is independent of database management systems (DBMS), we were able to continue using the PostgreSQL previously constructed. PostgreSQL is a fully functional open source DBMS with over 15 years of reliable operation and many sophisticated features. It is continually upgraded and its SQL implementation strongly conforms to the ANSI-SQL 92/99 standards. The Chado schema is well integrated with Gbrowse, the genome viewer available from GMOD (see below).
Currently, the annotations for the predicted genes in the purple sea urchin genome are organized into a separate PostgreSQL database using a simple schema derived from the original annotation fields used at BCM-HGSC. This allowed a rapid and relatively error-free transfer of the data to SpBase. Gene nomenclature, sequence coordinates and expression information are all included in this database. Editing and new entry are accomplished through a series of forms pages. Searching is available through a simple form with the most central attribute fields from the annotations. In addition, the gene information page offers links out to the two genome-wide expression studies on the sea urchin genome (16,19). Although expressed sequences were used in gene model predictions, these links provide additional information as to temporal patterns of expression.
For sequence searches we have incorporated the NCBI standalone web BLAST package [author: Sergei Shavirin, NCBI; (23)]. We offer sequence searches of both assemblies (0.5 and 2.1); the predicted gene models as nucleic acid sequence and protein sequence; and the sequences of the available clones from our genome facility arrayed libraries. This last category includes some individual BAC sequences, ESTs and many BAC-end sequences. In this arrangement, it is possible to ‘clone by blast’, e.g. obtain DNA clones from sequences identified experimentally. The blast results are linked out to sequence objects in the genome browser from which the actual sequence objects can be obtained.
We have chosen to use Gbrowse, the most popular genome browser included in the GMOD suite of software. It is richly endowed with many features for displaying sequences and their annotations (24). The constructor can include a variety of premade glyphs to document sequence features or design one's own. It is easy to attach arbitrary URLs to any annotation and thus link out to additional information. Furthermore, it supports third party annotations via GFF file formats. One can search on several different sequence attributes and easily download sequences. We offer either of the sequence assemblies in a Gbrowse format (Figure 2).
Figure 2.
A screen shot of the Gbrowse window for a specific gene model. The BACs and ESTs mapping to the scaffold from which the model originated are displayed. In addition to the sequences, the viewer has access to the actual clone designations and can order them from our facility.
While the sequence information is the central focus of this information system, we have also included several accessory functions as well. We have mounted a community bulletin board to facilitate discussion pertinent to the sea urchin genome information. In addition, a number of different text resources originally crafted for the SUGP web site have been copied over to SpBase. These include the annotated BAC maps and sequences developed as part of the Endomesoderm Gene Regulatory Network (25–27). A variety of methods specific to arrayed library preparation are posted in this section. A list of PCR primers used by sea urchin experimentalists is also displayed here. A space for supplemental data from genomics papers published by the sea urchin community is also maintained in this section.
THE FUTURE
The primary effort in these first months of the construction of SpBase has centered on the display of the existing genomic information for the purple sea urchin and comparative sequence information from other echinoderms. The majority of the sequences are those produced by BCM-HGSC and accessioned into GenBank. Almost all of the annotation information came directly from the databases at the BCM-HGSC web site. We have extensively edited these annotations to remove inconsistencies in, for example, gene names and to add missing information where available.
The primary change anticipated in the near future is the movement to a maintenance phase since the transfer is now complete. Most of the planned components are in place. We plan to add a Biomart search engine to support broad data mining functions. Other than that, our primary effort will be the collection and incorporation of new gene and genome data as available from literature sources as well as any future sequencing and assembly work. We expect to add a literature-based curation function such as Texpresso (28) to formalize the incorporation of new data.
FUNDING
National Institutes of Health; National Institute of Child Health and Human Development (R01HD056016); National Center for Research Resources; Division of Comparative Medicine (RR015044) and the Beckman Institute. Funding for open access charge: the US National Institute of Child Health and Human Development (R01HD056016).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Barbara Richer for carefully reading of the article. An earlier vesion of this information system was constructed with the aid of Emmanuelle Morin and Kris Khamvongsa.
REFERENCES
- 1.The Sea Urchin Sequencing Consortium. The purple sea urchin genome. Science. 2006;314:941–952. [Google Scholar]
- 2.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc. Natl Acad. Sci. USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodergren E, Shen Y, Song X, Zhang L, Gibbs R, Weinstock G. Shedding genetic light on Aristotle's lantern. Dev. Biol. 2006;300:2–8. doi: 10.1016/j.ydbio.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Springer MS, Tusneem NA, Davidson EH, Britten RJ. Phylogeny, rates of evolution, and patterns of codon usage among sea urchin retroviral-like elements, with implications for the recognition of horizontal transfer. Mol. Biol. Evol. 1995;12:219–230. doi: 10.1093/oxfordjournals.molbev.a040196. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez P, Lessios HA. Evolution of Sea Urchin Retroviral-Like (SURL) elements: evidence from 40 Echinoid species. Mol. Biol. Evol. 1999;16:938–952. doi: 10.1093/oxfordjournals.molbev.a026183. [DOI] [PubMed] [Google Scholar]
- 6.Littlewood DTJ, Smith AB. A combined morphological and molecular phylogeny for sea urchins (Echinoidea: Echinodermata) Phil. Trans. R. Soc. Lond. B. 1995;347:213–234. doi: 10.1098/rstb.1995.0023. [DOI] [PubMed] [Google Scholar]
- 7.Mooi R, Davids B. Skeletal homologies in Echinoderms. Paleontological Society Papers. 1997;3:305–335. [Google Scholar]
- 8.Cameron RA, Mahairas G, Rast JP, Martinez P, Biondi TR, Swartzell S, Wallace JC, Poustka AJ, Livingston BT, Wray GA, et al. A sea urchin genome project: sequence scan, virtual map, and additional resources. Proc. Natl Acad. Sci. USA. 2000;97:9514–9518. doi: 10.1073/pnas.160261897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havlak P, Chen R, Durbin KJ, Egan A, Ren Y, Song X-Z, Weinstock GM, Gibbs RA. The atlas genome assembly system. Genome Res. 2004;14:721–732. doi: 10.1101/gr.2264004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 11.Cai WW, Chen R, Gibbs RA, Bradley A. A clone-array pooled shotgun strategy for sequencing large genomes. Genome Res. 2001;11:1619–1623. doi: 10.1101/gr.198101. [DOI] [PubMed] [Google Scholar]
- 12.Poustka AJ, Groth D, Hennig S, Thamm S, Cameron RA, Beck A, Reinhardt R, Herwig R, Panopoulou G, Lehrach H. Generation, annotation, evolutionary analysis and database integration of 20,000 unique sea urchin EST clusters. Genome Res. 2003;13:2736–2746. doi: 10.1101/gr.1674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souvorov A, Tatusova T, Lipman DJ. Genome annotation with Gnomon—A multi-step combined gene prediction tool. National Center for Biotechnology Information. 2004 http://www.ncbi.nlm.nih.gov/projects/genome/guide/gnomon.shtml (30 October 2008, date last accessed) [Google Scholar]
- 14.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solovyev VV. Statistical approaches in eukaryotic gene prediction. In: Balding DEA, editor. Handbook of Statistical Genetics. Chichester, New York: John Wiley and Sons, Ltd; 2001. pp. 83–127. [Google Scholar]
- 16.Wei Z, Angerer RC, Angerer LM. A database of mRNA expression patterns for the sea urchin embryo. Dev. Biol. 2006;300:476–484. doi: 10.1016/j.ydbio.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SMJ, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsik CG, Worley KC, Zhang L, Milshina NV, Jiang H, Reese JT, Childs KL, Venkatraman A, Dickens CM, Weinstock GM, et al. Community annotation: procedures, protocols, and supporting tools. Genome Res. 2006;16:1329–1333. doi: 10.1101/gr.5580606. [DOI] [PubMed] [Google Scholar]
- 19.Samanta MP, Tongprasit W, Istrail S, Cameron RA, Tu Q, Davidson EH, Stolc V. The transcriptome of the sea urchin embryo. Science. 2006;314:960–962. doi: 10.1126/science.1131898. [DOI] [PubMed] [Google Scholar]
- 20.Howard-Ashby M, Materna SC, Brown CT, Tu Q, Oliveri P, Cameron RA, Davidson EH. High regulatory gene use in sea urchin embryogenesis: implications for bilaterian development and evolution. Dev. Biol. 2006;300:27–34. doi: 10.1016/j.ydbio.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:I337–I346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 22.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, et al. The future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C.-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev. Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- 26.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C.-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 27.Oliveri P, Davidson EH. Built to run, not fail. Science. 2007;315:1510–1511. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- 28.Müller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [Epub ahead of print 21 September 2004] [DOI] [PMC free article] [PubMed] [Google Scholar]