Abstract
The packing of protein atoms is an indicator for their stability and functionality, and applied in determining thermostability, in protein design, ligand binding and to identify flexible regions in proteins. Here, we present Voronoia, a database of atomic-scale packing data for protein 3D structures. It is based on an improved Voronoi Cell algorithm using hyperboloid interfaces to construct atomic volumes, and to resolve solvent-accessible and -inaccessible regions of atoms. The database contains atomic volumes, local packing densities and interior cavities calculated for 61 318 biological units from the PDB. A report for each structure summarizes the packing by residue and atom types, and lists the environment of interior cavities. The packing data are compared to a nonredundant set of structures from SCOP superfamilies. Both packing densities and cavities can be visualized in the 3D structures by the Jmol plugin. Additionally, PDB files can be submitted to the Voronoia server for calculation. This service performs calculations for most full-atomic protein structures within a few minutes. For batch jobs, a standalone version of the program with an optional PyMOL plugin is available for download. The database can be freely accessed at: http://bioinformatics.charite.de/voronoia.
INTRODUCTION
The packing of protein atoms is an important indicator for their stability and functionality. Measuring the packing of protein structures has been successfully applied to calculate the intrinsic compressibility of proteins (1,2), ligand binding and protein design (3), to find structural determinants for thermostability (4) and to identify flexible regions in proteins (5). Protein packing has recently received importance in assessing the quality of models in tertiary structure prediction (6), and to find water molecules not resolved in the crystal structure. However, tools for the analysis of packing are sparse. Here, we present Voronoia, a database for atomic-scale packing density analysis, also available as a downloadable application.
Proteins are not homogeneously packed; in their interior, they contain many atom-sized cavities that preferably locate close to the surface (7). Often, water molecules that reside in these cavities are not detected in crystal structures. We have observed an accumulation of cavities at functionally important sites of membrane channels and transporters (2,8). This conforms to an earlier finding that cavities are crucial for the conformational flexibility of photosynthetic proteins (9). Knowing the position of cavities allows to predict the effects of mutations (10), and thus to engineer proteins with increased stability (11). In neurotransmitter receptors, interior cavities are believed to form the site of action for some anesthetics (12).
Several procedures to assess the spatial proximity of atoms in 3D space have been developed, most allocating space to Voronoi polyhedra (13). Atom radii were introduced to apply the Voronoi procedure to proteins (14). This method was further improved by the Voronoi Cell method, using curved instead of planar interfaces and treating atoms at cavities and at the protein surface separately (15). An alternative approach is the Alpha Shape (AS) method (16) which models atoms as self-inflating spheres. Mathematically, this is a generalization of the Voronoi method applicable to a multitude of domains other than biomolecules, but it lacks the ability to calculate packing densities for atoms exposed to the solvent. Finally, the Occluded Surface (OS) method (17) calculates the length of rays protruding from an atom, but overestimates densities for atoms adjacent to cavities (2), making it more difficult to detect subtle packing differences in these regions. The AS and OS methods are available as downloadable programs. While these methods produce reasonable data, there are only few possibilities to report and visualize the results, especially for nonprogrammers. Other software comes with comfortable user interfaces, but has limited usage: SURFNET (18), CastP (19) and CAVER (20) calculate cavities and surface pockets in detail but no packing. Voro3D offers a wide range of functions but is restricted to residue-wise calculation of packing (21).
The Voronoi Cell method
These limitations are overcome by Voronoia, an implementation of the state-of-the-art Voronoi Cell algorithm (15). It includes construction of hyperboloid instead of planar interfaces, assigns individual radii for each atom, and separates the atomic volume between a Van der Waals sphere and a solvent-excluded portion. While the allocation of space is similar to that made by the AS method, the definition of a solvent excluded volume provides an extra level of detail, especially in protein regions with large packing defects. The volumes are calculated numerically by applying a cubic lattice to the protein and calculating each cubiculum separately. This approach reduces computation time to a few minutes for most structures. The resulting per-atom measure is the packing density:
where Vvdw is the assigned atomic volume inside the atoms’ Van der Waals radius and Vse is the remaining solvent excluded volume. The Voronoi Cell procedure uses the empirical ProtOr (22) set of atom radii, and distinguishes between buried atoms, surface atoms and atoms neighboring internal cavities.
Voronoia web interface
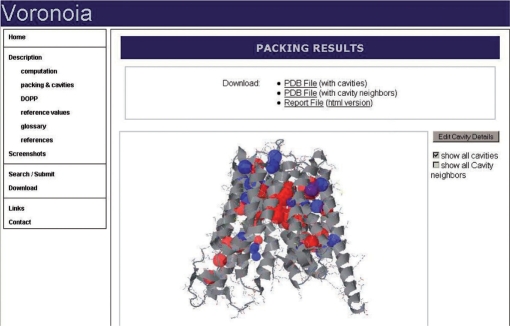
The calculation of packing files at higher accuracy is a time-consuming process. To allow the user to quickly visualize the packing of a particular PDB structure, we created an online database of packing data for all biological units from the PDB using the Voronoia program. In total, 61 318 pre-calculated packing files calculated with a grid distance of 0.02 Å, the highest accuracy possible. The results may be displayed in textual form, summarizing packing densities for buried atoms in the structure, and for each residue separately. Also, a complete list of interior cavities and their environment is given. For visual inspection, a Jmol-based 3D plugin allows to distinguish tightly and loosely packed regions in a structure, and to locate interior cavities (Figure 1). To query the database, a live search of PDB headers is enabled by an AJAX-based mechanism.
Figure 1.
Screenshot of the 3D visualization of an entry in Voronoia. Here, the glycerol-3-phosphate transporter (PDB code 1pw4) is shown. It has a transmembrane domain, in which a number of nonpolar (blue) and polar (red) cavities indicate regions of enhanced flexibility.
Reference packing data calculated from a current nonredundant set of 744 structures based on SCOP superfamilies is provided. For each structure in the database, its deviation from the reference values is given as a z-score RMS. If a particular file cannot be calculated, e.g. because the source data violates the PDB format or the file size reaches hundreds of MB, the user will be informed. This database will be updated every 6 months by means of an automatic script. A web interface has been set up at that allows performing the packing analysis for arbitrary input structures. The Voronoia web service performs calculations for most full-atomic protein structures within one to a few minutes. A lattice resolution for the Voronoi Cell algorithm of up to 0.1 Å is available on the web server, which is sufficient for most applications.
A standalone version of the Voronoia program is available with and without graphical interface. It uses regular PDB structure files as input, and creates modified PDB files containing packing densities for each atom. Both cavities and their surroundings may be written as separate structure files. Textual reports that summarize all this information and the deviation of packing from reference data are created for each structure. Voronoia is capable of creating these reports for a batch of structures in one go. This also generates average packing values, which may be used as a new reference set. Parameters such as the resolution of the cubic lattice, and the subset of atoms considered may be specified. Options going beyond those of the web database include choosing atom radii between the empirical ProtOr set (22) and a set optimized for molecular dynamics (23). Using Voronoia as a plugin for the PyMOL molecular viewer, the packing density, its deviation from reference values and cavities can be visualized immediately upon calculation.
CONCLUSION
Concluding, Voronoia is the first comprehensive easy-to-use toolkit for packing analysis. It allows browsing of pre-calculated packing densities and cavities, and de novo calculation for single protein structures and data sets, making it useful for a wide range of applications. Voronoia is available at http://bioinformatics.charite.de/voronoia.
FUNDING
European Union 6th Framework Programme Marie Curie Research Training Network ‘DNA Enzymes’ (MRTNCT-2005-019566); Deutsche Forschungsgemeinschaft (740 and 449). Funding for open access charges: Deutsche Forschungsgemeinschaft (SFB-449).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Cornelius Frömmel for initiating this packing project and kind support. We also thank Fiete Haak for implementing the initial version of the website.
REFERENCES
- 1.Paci E, Marchi M. Intrinsic compressibility and volume compression in solvated proteins by molecular dynamics simulation at high pressure. Proc. Natl Acad. Sci. USA. 1996;93:11609–11614. doi: 10.1073/pnas.93.21.11609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hildebrand PW, Rother K, Goede A, Preissner R, Frömmel C. Molecular packing and packing defects in helical membrane proteins. Biophys. J. 2005;88:1970–1977. doi: 10.1529/biophysj.104.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glyakina AV, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. Different packing of external residues can explain differences in the thermostability of proteins from thermophililic and mesophilic organisms. Bioinformatics. 2007;23:2231–2238. doi: 10.1093/bioinformatics/btm345. [DOI] [PubMed] [Google Scholar]
- 5.Jernigan RL, Kloczkowski A. Packing regularities in biological structures relate to their dynamics. Methods Mol. Biol. 2007;350:251–276. doi: 10.1385/1-59745-189-4:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pawlowski M, Gajda MJ, Matlak R, Bujnicki JM. MetaMQAP: a meta-server for the quality assessment of protein models. BMC Bioinformatics. 2008;9:403. doi: 10.1186/1471-2105-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rother K, Preissner R, Goede A, Frömmel C. Inhomogeneous molecular density: reference packing densities and distribution of cavities within proteins. Bioinformatics. 2003;19:2112–2121. doi: 10.1093/bioinformatics/btg292. [DOI] [PubMed] [Google Scholar]
- 8.Hildebrand PW, Gunther S, Goede A, Forrest L, Frommel C, Preissner R. Hydrogen-bonding and packing features of membrane proteins: functional implications. Biophys. J. 2008;94:1945–1953. doi: 10.1529/biophysj.107.110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shlyk-Kerner O, Samish I, Kaftan D, Holland N, Sai PS, Kless H, Scherz A. Protein flexibility acclimatizes photosynthetic energy conversion to the ambient temperature. Nature. 2006;442:827–830. doi: 10.1038/nature04947. [DOI] [PubMed] [Google Scholar]
- 10.Cuff AL, Martin AC. Analysis of void volumes in proteins and application to stability of the p53 tumour suppressor protein. J. Mol. Biol. 2004;344:1199–1209. doi: 10.1016/j.jmb.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Teerinen T, Valjakka J, Rouvinen J, Takkinen K. Structure-based stability engineering of the mouse IgG1 Fab fragment by modifying constant domains. J. Mol. Biol. 2006;361:687–697. doi: 10.1016/j.jmb.2006.06.073. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Loll PJ, Eckenhoff RG. Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein. FASEB J. 2005;19:567–576. doi: 10.1096/fj.04-3171com. [DOI] [PubMed] [Google Scholar]
- 13.Voronoi GF. Nouvelles applications des paramètres continus à la théorie de formes quadratiques. J. angew. reine Math. 1908;134:198–287. [Google Scholar]
- 14.Richards FM. The interpretation of protein structures: total volume, group volume distributions and packing density. J. Mol. Biol. 1974;82:1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- 15.Goede A, Preissner R, Frömmel C. Voronoi Cell: new method for allocation of space among atoms: elimination of avoidable errors in calculation of atomic volume and density. J. Comp. Chem. 1997;18:1113–1123. [Google Scholar]
- 16.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: I. Molecular area and volume through alpha shape. Proteins. 1998;33:1–17. [PubMed] [Google Scholar]
- 17.Pattabiraman N, Ward KB, Fleming PJ. Occluded molecular surface: analysis of protein packing. J. Mol. Recognit. 1995;8:334–344. doi: 10.1002/jmr.300080603. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities and intermolecular interactions. J. Mol. Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 19.Binkowski TA, Naghibzadeh S, Liang J. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrek M, Otyepka M, Banás P, Kosinová P, Koca J, Damborský J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupuis F, Sadoc JF, Jullien R, Angelov B, Mornon JP. Voro3D: 3D Voronoi tessellations applied to protein structures. Bioinformatics. 2005;21:1715–1716. doi: 10.1093/bioinformatics/bth365. [DOI] [PubMed] [Google Scholar]
- 22.Tsai J, Taylor R, Chothia C, Gerstein M. The packing density in proteins: standard radii and volumes. J. Mol. Biol. 1999;290:253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 23.Stouten PFW, Frömmel C, Nakamura H, Sander C. An effective solvation term based an a atomic occupancies for use in protein simulations. Mol. Simulat. 1993;10:97–120. [Google Scholar]