Abstract
McKusick's Online Mendelian Inheritance in Man (OMIM®; http://www.ncbi.nlm.nih.gov/omim), a knowledgebase of human genes and phenotypes, was originally published as a book, Mendelian Inheritance in Man, in 1966. The content of OMIM is derived exclusively from the published biomedical literature and is updated daily. It currently contains 18 961 full-text entries describing phenotypes and genes. To date, 2239 genes have mutations causing disease, and 3770 diseases have a molecular basis. Approximately 70 new entries are added and 700 entries are updated per month. OMIM® is expanding content and organization in response to shifting biological paradigms and advancing biotechnology.
INTRODUCTION
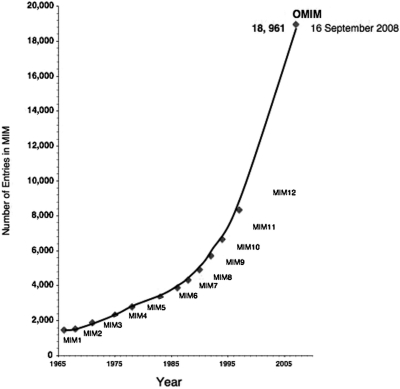
Dr Victor A. McKusick wrote an article in 1962 for the Quarterly Review of Biology titled ‘On the X Chromosome of Man’ (1). At that time, X-linkage had been established for about 60 traits in man and a genetic map of the X chromosome was presented. Four years later, with the addition of dominant and recessive traits, Dr McKusick published a book, Mendelian Inheritance in Man: Catalogs of Autosomal Dominant, Autosomal Recessive and X-linked Phenotypes (MIM) (2). The first edition had 1400 entries and no mapped autosomal loci. Each catalogue contained summaries of genetic phenotypes reported in the biomedical literature, which were organized into numbered entries with descriptive synopses and references. With the addition of descriptions of genes, the focus of MIM became the relationship between phenotypes and genes. Over the years, catalogues for Y-linked and mitochondrial phenotypes and genes were added and by the 12th edition, the subtitle had been changed to ‘A Catalog of Human Genes and Genetic Disorders’(3). Today the online version of MIM, OMIM®, contains 18 961 entries (Figure 1). It continues with the same basic organization but has grown to include complex traits and descriptions of the consequences of gene copy number variation and recurrent deletions/microdeletions and duplications/microduplications. A general overview on how to search OMIM® was described by Hamosh et al. (4). This paper describes the content and organization of OMIM® and the modifications that have been made to accommodate advances in our understanding of genomic architecture and complex traits.
Figure 1.
Number of entries in Mendelian Inheritance in Man.
DATABASE CONTENT AND ORGANIZATION
The content of OMIM® continues to be based on the peer-reviewed biomedical literature. Journals are scanned every day for new information on Mendelian disorders and genes already in the database as well as newly described genes and disorders. Articles are then selected for review and possible inclusion with priority given to papers describing genes associated with disease phenotypes, genes with substantial new biology and disorders and genes not in OMIM®. In addition, other online genetics resources are scanned for information and articles that may be relevant to OMIM®. As can be seen from the ‘update log’ on the blue bar of the OMIM® Home Page, approximately 70 new entries are created and 700 existing entries are updated per month.
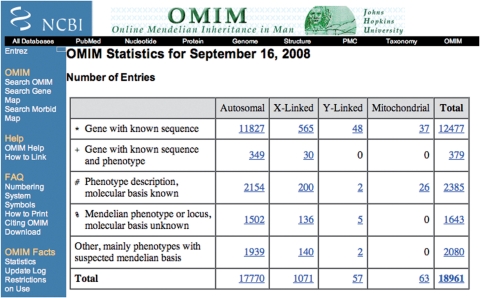
Each OMIM® entry is given a unique six digit number. The symbol (*, #,% or ⁁) that precedes the number distinguishes between phenotype entries and gene entries. As of 16 September 2008, OMIM® contains 18 961 entries (Figure 2). Of these, 6487 entries contain descriptions of phenotypes. As always, the primary focus of OMIM® is Mendelian phenotypes and their genes. With the completion of the sequencing of the human genome, and the proliferation of cloned genes, OMIM® has directed its attention to genes with direct disease relationship and those of known function. OMIM® currently contains 12 856 gene entries.
Figure 2.
OMIM® statistics.
The relationship between phenotype and genotype can be complex. At the most simple level, when the molecular basis is known for a phenotype, a number sign (#) is added to the MIM number of the phenotype entry and allelic variants (mutations) are added to the gene entry. The first paragraph of the phenotype entry will always state why a number sign is used. Some entries with a plus sign (+) include both phenotype and gene information and are awaiting review and separation. An allelic variant is designated by the MIM number of its parent entry, followed by a decimal point and a unique 4-digit variant number. For example, allelic variants in the cystic fibrosis transmembrane conductance regulator gene (CFTR, MIM 602521) are numbered 602 421.0001 to 602 421.0136. The β-globin locus (HBB) is numbered 141900; sickle haemoglobin (HbS) is numbered 141900.0243. Descriptions of sickle cell anaemia and cystic fibrosis are contained in entries 603903 and 219700, respectively. For most genes, only selected mutations are included as allelic variants. Criteria for inclusion are the first mutation to be discovered, distinctive phenotype, high population frequency, historic significance, unusual mechanism of mutation, unusual pathogenetic mechanism and distinctive inheritance (e.g. dominant with some mutations, recessive with other mutations in the same gene, as in the case of mutations in connexin 26 (MIM 121011), which can cause recessive and dominant forms of deafness, among other phenotypes). Most of the allelic variants represent disease-producing mutations. A few polymorphisms are included, many of which show a positive statistical correlation with particular common disorders. A small but increasing number of allelic variants have been linked to dbSNP. These links, when available, appear after the mutation listed in each allelic variant. A list of the 2239 gene entries in OMIM® with disease-causing mutations can be obtained by searching on 0001 with the LIMITS restricted to ‘Allelic Variants’.
Because OMIM® does not catalogue all allelic variants in any gene, it includes links to several complementary resources. These include the Human Gene Mutation Database (HGMD, www.hgmd.cf.ac.uk/), a repository for published mutations, and over 500 locus-specific mutation databases through the Human Genome Variation Society (HGVS, www.hgvs.org/dblist/dblist/html). These links are available from the blue bar to the left of the screen when viewing an OMIM® entry.
In the early days of OMIM®, information added to an existing entry was placed at the end of the text to reflect the historical development of knowledge of the phenotype or gene. As the knowledge of genes and phenotypes expanded and became more detailed, greater structure was added to the OMIM® entries. Headings and subheadings have been added to organize related information while retaining historical development (Table 1). Free-text within these headings and subheadings allows discussion of the nuances of genotype/phenotype correlation that is not possible when bound by more rigid data structures. For example, the complex subject of imprinting that occurs in genes and results in variable phenotypes can be discussed in separate ‘imprinting’ headings in the gene and the related phenotype entry, thus providing complementary perspectives on the subject.
Table 1.
Major OMIM Text headings
| Phenotype entry |
| • Description |
| • Clinical features |
| • Biochemical features |
| • Inheritance |
| • Mapping |
| • Pathogenesis |
| • Diagnosis |
| • Genotype/phenotype correlations |
| • Clinical management |
| • Population genetics |
| • Molecular genetics |
| • Animal model |
| • History |
| Gene entry |
| • Description |
| • Cloning |
| • Gene structure |
| • Gene function |
| • Evolution |
| • Mapping |
| • Molecular genetics |
| • Genotype/phenotype correlations |
| • Animal model |
| • History |
| • Allelic Variantsa |
An OMIM entry contains headings based on current knowledge of the topic. Text headings can contain subheadings to provide additional organization to each entry.
aAllelic Variants, a section found only in gene entries, is a field heading in the same hierarchy as Number, Title, Text and References.
In general, OMIM® splits genetically heterogeneous phenotypes. This has proven to be a good strategy, as the different long QT syndromes (LQT1-10), initially clinically indistinguishable, have now been shown to be caused by mutations in different ion channel genes. This improved understanding of pathophysiology has led to individualized clinical management of the disease. The first member of a phenotypic series (e.g. LQT1, MIM 192500) will usually include the overview information on the group of disorders.
THE MORBID AND SYNOPSIS MAPS
The Morbid Map and the Synopsis Map provide a shorthand view of the relationship between disease and gene. The disorders with a ‘(3)’ after them represent phenotypes for which the molecular basis is known. Currently, there are 3770 diseases or disease susceptibilities that have an underlying molecular basis. Clicking on the cytogenetic location in either the Morbid or Synopsis Map reveals a more detailed, sequence-based view of the latest genome build and transcripts within that cytogenetic region. The Morbid Map data are included in NCBI's Entrez Gene, and users are encouraged to use MapViewer directly.
CLINICAL SYNOPSES
Over 4500 phenotype entries have associated clinical synopses. Over 1500 of these have been revised or are newly written. Each feature in a new or revised synopsis is derived from the published literature and standard clinical references. The structure of the clinical synopses is based on an anatomic template, beginning with Inheritance and ending with Molecular Basis, when known. OMIM® uses regular, but not controlled vocabulary and often defers to the author's original description. OMIM® clinical feature terms are incorporated into the National Library of Medicine, UMLS project (Unified Medical Language System) and SNOMED (Systematized Nomenclature of Medicine).
NEW OMIM® DEVELOPMENTS
Because OMIM® is a free-text database, it can easily accommodate descriptions of paradigm shifts in biology. OMIM® now includes descriptions of microRNAs, non-coding regulatory elements and modifier genes.
Contiguous gene syndromes such as Williams syndrome and Miller-Dieker syndrome have long been included in OMIM®. The advent of microarray technology has enhanced the ability to detect and characterize these chromosomal duplication and deletion syndromes. OMIM® includes such chromosomal aberrations when the genes identified in the region can be related to the phenotype at the gene function level. The cytogenetic location of the duplication/deletion is included in the title [e.g. chromosome 11p13 deletion syndrome (MIM 194072), chromosome 10q26 deletion syndrome (MIM 609625)].
It is now known that some genes occur in multiple copy number that varies among individuals (e.g. CCL3L1, β-defensin, amylase 1). Variation in copy number of these genes may have phenotypic consequences. For example, variation in CCL3L1 (MIM 601395) copy number is known to influence immune reconstitution after antiretroviral therapy for HIV infection. When the copy number variation has recognized relevance to human disease, this is addressed in both the phenotype and the gene entry.
Genome-wide association (GWA) studies of complex traits represent the next step in genetic mapping of phenotypes. GWA studies implicate numerous chromosomal regions, but the findings are often not replicated. To limit as much as possible the creation of invalid phenotypic loci, OMIM® has established criteria for the inclusion of complex trait loci based on such studies. These criteria include observation of similar results in at least three independent samples, P-values of <10−7 after Bonferroni correction, effect size of at least 1.2-fold relative risk or odds ratio, and characterization of the functional effect. When the criteria have been met, a phenotypic series is created [e.g. inflammatory bowel disease (IBD) 1–20]. The series can be retrieved by typing the root symbol followed by an asterisk; in this case IBD*. Information from GWA studies will often be put into the Mapping heading of an entry, unless a functional relationship has been shown. In that case, the information will be put in as an Allelic Variant of the gene.
Many genetic traits are quantifiable and are related to ‘normal’ ranges in human anatomy and physiology. To address the spectrum in these phenotypes, OMIM® is creating phenotypic quantitative trait loci (QTL) series (e.g., body mass index, cholesterol, bone mineral density and blood pressure). Discussions of the disease states of these QTLs were already in OMIM® (obesity/leanness, hypercholesterolemia, osteoporosis/osteopetrosis and hypertension, respectively). Bringing together these phenotypic spectra under the QTL heading is an ongoing challenge.
LINKING TO AND FROM OMIM®
OMIM® numbers are stable, cross-referenced and frequently included in many publications. Other databases are encouraged to include OMIM® numbers in their datasets. This will facilitate linking to and from OMIM® (see ‘How to Link’ in the blue bar on the left of the OMIM® home page). Additionally, the text of OMIM® and the Morbid Map are available for download subject to some restrictions (see ‘Restrictions on Use’ in the blue bar). Finally, NCBI has created some useful web and programming tools for data retrieval from OMIM® and other Entrez databases (http://eutils.ncbi.nlm.nih.gov/entrez/query/static/advancedentrez.html), and a file cross-referencing OMIM® numbers to gene ID numbers is maintained for ftp download (ftp.ncbi.nlm.nih.gov/gene/DATA/mim2gene).
OMIM® welcomes collaboration and users are encouraged to submit comments and corrections to info@ncbi.nlm.nih.gov. All suggestions must be accompanied by a full supporting citation.
FUNDING
The National Human Genome Research Institute (to OMIM®); the National Library of Medicine (to OMIM®). Funding for open access charge: N01LM43504.
Conflict of interest statement. None declared.
REFERENCES
- 1.McKusick VA. On the X Chromosome of Man. Quart. Rev. Biol. 1962;37:69–175. doi: 10.1086/403631. [DOI] [PubMed] [Google Scholar]
- 2.McKusick VA. Mendelian Inheritance in Man, A Catolog of Autosomal Dominant, Autosomal Recessive, and X-linked Phenotypes. 1st edn. Baltimore, MD: Johns Hopkins University Press; 1966. [Google Scholar]
- 3.McKusick VA. Mendelian Inheritance in Man, A Catolog of Human Genes and Genetic Disorders. 12th edn. Baltimore, MD: Johns Hopkins University Press; 1998. [Google Scholar]
- 4.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM®), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:514–517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]