Abstract
We have developed the database TMFunction, which is a collection of more than 2900 experimentally observed functional residues in membrane proteins. Each entry includes the numerical values for the parameters IC50 (measure of the effectiveness of a compound in inhibiting biological function), Vmax (maximal velocity of transport), relative activity of mutants with respect to wild-type protein, binding affinity, dissociation constant, etc., which are important for understanding the sequence–structure–function relationship of membrane proteins. In addition, we have provided information about name and source of the protein, Uniprot and Protein Data Bank codes, mutational and literature information. Furthermore, TMFunction is linked to related databases and other resources. We have set up a web interface with different search and display options so that users have the ability to get the data in several ways. TMFunction is freely available at http://tmbeta-genome.cbrc.jp/TMFunction/.
INTRODUCTION
Membrane proteins perform a diverse variety of functions and are used as main drug targets of pharmaceutical agents. The collection of information on potential amino acid residues for the function of membrane proteins is important for understanding the sequence–structure–function relationship of membrane proteins as well as predicting the functional residues from sequence/structure. Tusnady et al. (1) constructed a database for transmembrane proteins, which covers the three-dimensional structures of membrane proteins deposited in Protein Data Bank (PDB) and information on membrane spanning α-helices and β-strands obtained with the TMDET algorithm (2). Saier et al. (3) developed a comprehensive classification system for membrane transport proteins known as the Transport Classification Database (TCDB). Further, functional databases have been developed for G-protein-coupled receptors (GPGRs), human seven transmembrane receptors, Arabidopsis integral membrane proteins and so on (4,5). Edvardsen et al. (6) created a G-protein-coupled receptor mutant database, which is mainly focused on different families of GPCRs. Mutational databases have also been developed for the structure, function and thermodynamics of proteins (7,8). On the other hand, several methods have been proposed for discriminating transmembrane α-helical and β-strand proteins, predicting their membrane-spanning segments, and functional classification of membrane proteins (9–16). In spite of these studies, there is no database available for the broad collection of potential residues, which are important for the functions of different classes of membrane proteins including receptors, transporters, channel proteins, etc. This information can be obtained from experimental studies on membrane proteins that have reported the measured values of several parameters for membrane protein function. The collection of such data is important and necessary for analyzing and predicting potentially important residues for practical applications.
In this work, we have developed a database, TMFunction, which is a collection of more than 2900 experimental data about important amino acid residues in membrane proteins, reported in the literature. It has information about functionally important residues, numerical values for the parameters IC50, Vmax, activity, affinity, etc., along with sequence and structure information for the protein, mutational and literature information. This database will help in understanding the relationship between amino acid sequences/structures and functions of membrane proteins. We have developed a WWW interface to facilitate searching the database and displaying the results with different options.
CONTENTS OF THE DATABASE
Each entry in the database includes the following information:
Sequence and structure information: name and source of the protein, Uniprot (17) and PDB (18) codes, type of the integral membrane protein (α-helical or β-strand), mutational details (single, double or wild-type; mutation has been identified with mutated residue, residue number and mutant residue; e.g. A102V) and location of mutants.
Functional information: factors that are mainly affected by the mutation of amino acid residues in membrane proteins, such as relative activity of mutants, affinity for binding, channel, drug, glycosylation, membrane insertion, cellular signaling, membrane translocation, transport, etc.
Functional data: numerical values for affinity (%), Bmax, IC50, drug sensitivity, Kd, Km, Vmax, uptake (%), etc.
Experimental methods and conditions: measurement, method and the ligand used for the control data.
Literature information: keywords, reference, authors and remarks.
DATABASE STATISTICS
The first release of TMFunction contains 2907 entries from 83 different proteins, which perform 29 diverse functions. It has data for 2092 single mutants, 580 multiple mutants and 273 wild-types. The majority of the data concerns transmembrane helical proteins (2760) followed by β-barrel membrane proteins (147). The data are obtained from more than 100 scientific articles published in 20 different journals.
FEATURES OF TMFUNCTION
TMFunction includes several features in the search and display options as shown in Figure 1, and as briefly explained subsequently:
Retrieving data for a particular protein and/or source.
Specifying the type of the mutant as single, multiple and/or wild-type.
Selecting the function of the protein (transport) as well as the numerical value of the functional parameter (e.g. IC50).
Mentioning the type of the protein.
Extracting data by authors, publication year and keywords.
Downloading entire data.
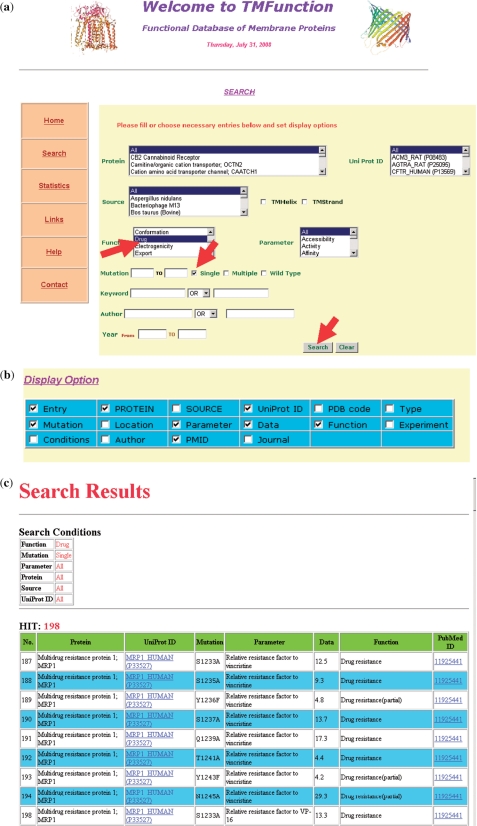
Figure 1.
An example of searching conditions, display options and results of TMFunction: (a) main menu for the search options in TMFunction. The items function (drug) and single mutants are selected for search as indicated by arrows; (b) display options in TMFunction. We have selected entry, protein, Uniprot ID, mutation, parameter, data, function and PMID to show in the output; (c) part of the results obtained from TMFunction.
Detailed tutorials describing the usage of TMFunction are available at the home page. For example, the data obtained for the function ‘drug’ and single mutants is shown in Figure 1a. The terms: entry, protein, Uniprot ID, mutation, parameter, data, function, experiments and Pubmed ID have been selected for displaying the results (Figure 1b). Figure 1c shows the final results obtained with the search conditions and display options.
LINKS TO OTHER DATABASES
Each entry in TMFunction is linked to Uniprot ID (http://www.uniprot.org/) and PDB code (http://www.rcsb.org) to obtain the sequence and structure information directly. The references for all data are directly connected to the PUBMED literature database of NCBI (http://www.ncbi.nlm.nih.gov/pubmed/). Further, we have provided links to several related databases and web servers including sequences and structures, functions and genomes, transmembrane helix and strand predictions (http://tmbeta-genome.cbrc.jp/TMFunction/DBlinkspage.html).
AVAILABILITY AND CITATION OF TMFUNCTION
The database can be freely accessible at http://tmbeta-genome.cbrc.jp/TMFunction/. If this database is used as a tool in your published research work, please cite this article including the URL. Suggestions and comments are welcome and should be sent to michael-gromiha@aist.go.jp.
FUNDING
The support from Computational Biology Research Center (CBRC) and National Institute of Advanced Industrial Science and Technology (AIST) is gratefully acknowledged. Funding for open access charge: Computational Biology Research Center (CBRC).
Conflict of interest statement: None declared.
ACKNOWLEDGEMENT
We thank Dr Martin Frith for critical reading of the manuscript.
REFERENCES
- 1.Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tusnady GE, Dosztanyi Z, Simon I. TMDET: web server for detecting transmembrane regions of proteins by using their 3D coordinates. Bioinformatics. 2005;21:1276–1277. doi: 10.1093/bioinformatics/bti121. [DOI] [PubMed] [Google Scholar]
- 3.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horn F, Bettler E, Oliveira L, Campagne F, Cohen FE, Vriend G. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwacke R, Schneider A, Van Der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edvardsen O, Reiersen AL, Beukers MW, Kristiansen K. tGRAP, the G-protein coupled receptors mutant database. Nucleic Acids Res. 2002;30:361–363. doi: 10.1093/nar/30.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawabata T, Ota M, Nishikawa K. The Protein Mutant Database. Nucleic Acids Res. 1999;27:355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gromiha MM, An J, Kono H, Oobatake M, Uedaira H, Sarai A. ProTherm: thermodynamic database for proteins and mutants. Nucleic Acids Res. 1999;27:286–288. doi: 10.1093/nar/27.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 10.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 11.Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 12.Lehnert U, Xia Y, Royce TE, Goh CS, Liu Y, Senes A, Yu H, Zhang ZL, Engelman DM, Gerstein M. Computational analysis of membrane proteins: genomic occurrence, structure prediction and helix interactions. Q. Rev. Biophys. 2004;37:121–146. doi: 10.1017/s003358350400397x. [DOI] [PubMed] [Google Scholar]
- 13.Gromiha MM, Suwa M. Current developments on beta-barrel membrane proteins: sequence and structure analysis, discrimination and prediction. Curr. Protein Pept. Sci. 2007;8:580–599. doi: 10.2174/138920307783018712. [DOI] [PubMed] [Google Scholar]
- 14.Gromiha MM, Yabuki Y, Kundu S, Suharnan S, Suwa M. TMBETA-GENOME: database for annotated beta-barrel membrane proteins in genomic sequences. Nucleic Acids Res. 2007;35:D314–D316. doi: 10.1093/nar/gkl805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromiha MM, Yabuki Y. Functional discrimination of membrane proteins using machine learning techniques. BMC Bioinform. 2008;9:135. doi: 10.1186/1471-2105-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Dai X, Zhao X. A nearest neighbor approach for automated transporter prediction and categorization from protein sequences. Bioinformatics. 2008;24:1129–1136. doi: 10.1093/bioinformatics/btn099. [DOI] [PubMed] [Google Scholar]
- 17.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]