Abstract
FlyBase (http://flybase.org) is a database of Drosophila genetic and genomic information. Gene Ontology (GO) terms are used to describe three attributes of wild-type gene products: their molecular function, the biological processes in which they play a role, and their subcellular location. This article describes recent changes to the FlyBase GO annotation strategy that are improving the quality of the GO annotation data. Many of these changes stem from our participation in the GO Reference Genome Annotation Project—a multi-database collaboration producing comprehensive GO annotation sets for 12 diverse species.
INTRODUCTION TO GENE ONTOLOGY ANNOTATION
What gene products do and where they do it are fundamental questions for biologists no matter what organism is being studied. The Gene Ontology (GO; www.geneontology.org) was established 10 years ago as a means of summarizing this information consistently across different databases by using a common set of defined controlled vocabulary terms. FlyBase was one of three founding members of the Gene Ontology Consortium [GOC; (1)] but the GO has since been adopted by many model organism databases, making comparison of gene function between diverse species feasible. The GO also encodes relationships between terms, which allows for efficient searching and computational reasoning. For example, a search for all gene products with ‘kinase activity’ will automatically gather those products labelled with the more specific types of kinase activity.
GO annotation comprises at least three components: a GO term that describes molecular function, biological role or subcellular location; an ‘evidence code’ that describes the type of analysis used to support the GO term (Table 1); and an attribution to a specific reference. There may also be supporting evidence for the choice of GO term in the form of database cross-references; for instance, a gene function may be ‘inferred from genetic interaction’, in which case an identifier for the interacting gene will be included. Qualifiers may also be included to modify the annotation; currently these are: ‘colocalizes_with’, ‘contributes_to’ and ‘NOT’. In the case of the ‘NOT’ qualifier, this has the effect of negating the annotation. FlyBase uses ‘NOT’ sparingly for cases where there is a prior expectation that a GO term should apply to a gene product but evidence exists to the contrary. Another annotation component, one that FlyBase has neglected until recently, is date. FlyBase now records an accurate date for each GO annotation reflecting when the annotation was originally made or last reviewed by a curator. GO annotations made prior to implementing the data component are dated 20060803. Examples of GO annotations are shown in Table 2.
Table 1.
Evidence codes used in GO annotation
| Manually assigned evidence codes |
| Experimental evidence codes |
| Inferred from Direct Assay (IDA) |
| Inferred from Physical Interaction (IPI) |
| Inferred from Mutant Phenotype (IMP) |
| Inferred from Genetic Interaction (IGI) |
| Inferred from Expression Pattern (IEP) |
| Computational analysis evidence codes |
| Inferred from Sequence or Structural Similarity (ISS) |
| Inferred from Sequence Orthology (ISO)a |
| Inferred from Sequence Alignment (ISA)a |
| Inferred from Sequence Model (ISM)a |
| Inferred from Genomic Context (IGC) |
| Inferred from Reviewed Computational Analysis (RCA) |
| Author statement evidence codes |
| Traceable Author Statement (TAS) |
| Non-traceable Author Statement (NAS) |
| Curatorial statement codes |
| Inferred by Curator (IC) |
| No biological Data available (ND) |
| Automatically assigned evidence codes |
| Inferred from Electronic Annotation (IEA) |
aThree new subcategories of the ISS evidence code used to assign GO terms based on sequence similarity. ISO is used when the similar sequences are considered to be orthologous. ISA is used where there is extensive sequence alignment but the sequences are not known to be orthologous. ISM is used when a sequence model has been generated from a set of related sequences, e.g. hidden Markov models for transmembrane regions. Full documentation of evidence codes together with how they are used in annotation can be found on the GO website (http://www.geneontology.org/GO.contents.doc.shtml).
Table 2.
Examples of GO annotations for D. melanogaster genes
| 2. Object ID | 3. Object symbol | 4. Qualifier | 5. GO ID | 6. DB:Reference | 7. Evidence Code | 8. With/From | 15. Date |
|---|---|---|---|---|---|---|---|
| FBgn0029891 | Pink1 | GO:0007005 | FB:FBrf0193630|PMID:16672980 | IGI | FB:FBgn0040491 | 20070523 | |
| FBgn0034879 | Rrp4 | GO:0006397 | FB:FBrf0105495 | ISS | SGD:S0001111 | 20060803 | |
| FBgn0020615 | SelD | NOT | GO:0004756 | FB:FBrf0099751|PMID:9398525 | IDA | 20060803 | |
| FBgn0010349 | Dhc64C | colocalizes_with | GO:0005739 | FB:FBrf0191163|PMID:16467387 | IDA | 20071221 | |
| Fbgn0033687 | CG8407 | GO:0007017 | FB:FBrf0174215 | IEA | InterPro:IPR001372 | 20080731 | |
| FBgn0036811 | MED11 | contributes_to | GO:0016455 | FB:FBrf0150795|PMID:12021283 | IC | GO:0000119 | 20070523 |
The column numbers are identical to those in the gene_association.fb file; the full file contains 15 columns of information.
GO annotation is useful for both small-scale and large-scale analyses. It can provide a first indication of the nature of a gene product and, in conjunction with evidence codes, point directly to papers with pertinent experimental data. However, it is particularly useful in the analysis of large data sets such as the output of a microarray experiment. For instance, the AmiGO GO Term Enrichment Tool (amigo.geneontology.org/cgi-bin/amigo/term_enrichment) will show significant shared GO terms or parents of those GO terms associated with a list of genes. Whatever the scale of use, for the best results it is important that GO annotation be as complete and accurate as possible. This article describes recent changes to GO annotation procedures in FlyBase that aim to improve the quality of our functional annotations.
GO ANNOTATION IN FLYBASE
GO annotations appear on the Gene Report page in FlyBase (Figure 1) and are also available as a downloadable file (gene_association.fb.gz in the genes section of the Precomputed Files page accessed from the Files menu on the FlyBase home page). This file is in the standard GO gene_association file format (www.geneontology.org/GO.format.annotation.shtml) and corresponds to the data that are submitted to the GOC for a given FlyBase release. GO data are searchable in FlyBase using both TermLink (retrieves data for all Drosophila species) and QueryBuilder (species and other criteria can be specified) (2).
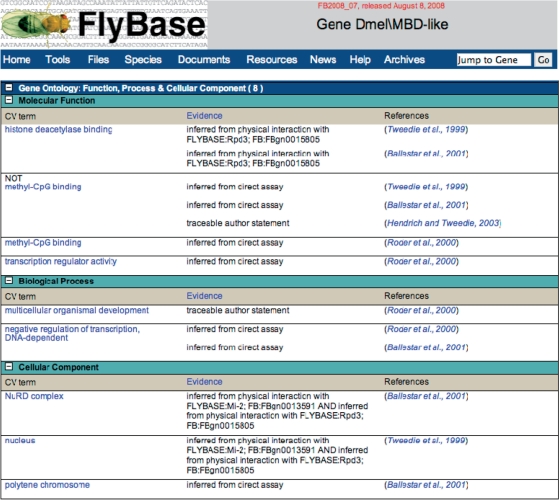
Figure 1.
GO annotation on the Gene Report of D. melanogaster MBD-like gene. Note the presence of contradictory experimental evidence for the term ‘methyl-CpG binding’ as indicated by the use of the qualifier ‘NOT’ for some publications. Each highlighted term links to a CV term report that includes a definition for the term and a diagram indicating its relationship to other terms.
Table 3 shows a summary of the current FlyBase GO annotations; 10 131 (72%) of the 14 029 Drosophila melanogaster protein-coding genes have at least one GO annotation and 9403 (67%) have annotations with specific (non-root) GO terms.
Table 3.
Numbers of D. melanogaster protein-coding genes (from a total of 14029 in FB2008_08) with GO annotation by evidence type and ontology
| Biological Process | Molecular Function | Cellular Component | Combined GO | |
|---|---|---|---|---|
| Genes with any GO annotation | 8080 | 9253 | 6893 | 10 131 |
| Genes with ≥1 experimentally based terma | 2603 | 1217 | 1403 | 3163 |
| Genes with only electronic annotationb | 2288 | 2055 | 1684 | 1716 |
| Genes with no data availablec | 855 | 859 | 1013 | 728 |
aAssigned with evidence codes: IDA, IPI, IMP, IGI, IEP.
bAssigned with IEA evidence code.
cRoot GO terms assigned with ND evidence code (note that ‘ND’ is applied only to genes that have been assessed for functional data; it is not used for genes that have not yet been subject to GO curation).
The GO is dynamic and its content can change on a daily basis. Most of these changes are the addition of new terms but other alterations, such as making a term obsolete, a term name change or a change to ontology structure, require us to revisit our existing annotations to choose alternative GO terms or check that the annotations are still valid. Rather than attempt to keep completely up-to-date with this moving target, FlyBase loads a new version of the GO every one or two releases of FlyBase. Both new and existing annotations are made consistent with this ‘frozen’ version for a given release. The frozen version of the GO, and all other ontologies used in FlyBase, are available in the Ontology Terms section of the Precomputed Files page.
The GO annotation set is submitted to the GOC at the same time as a new version of FlyBase is released. Users should be aware that there may be a few differences between the data downloadable from FlyBase and the file downloadable from the GO website. Differences arise because the submitted files are screened for errors, and lines that do not meet a series of quality controls are removed. For instance, any annotations with terms that have become obsolete since the ontology was frozen at FlyBase will be stripped out of the files available from the GO site.
GO CURATION PIPELINE
Most GO data in FlyBase are entered via our paper-by-paper literature curation process; curators read papers and chose the appropriate terms based on the data described. For gene products that have not yet been experimentally characterized, GO terms are assigned manually if there is significant sequence similarity to gene products of known function, using the ‘inferred from sequence similarity’ evidence code or one of the new more specific versions of this code (Table 1). When no specific term can be assigned from either published data or sequence homology, a gene is annotated with the non-specific ‘root’ term (molecular_function, biological_process or cellular_component) of the appropriate ontology, using the ‘no data available’ evidence code. This convention distinguishes between genes that are uncharacterized from those that lack GO terms because they have yet to be examined by a curator.
Manual curation is labour-intensive and it is a constant challenge to keep up to date with the volume of Drosophila literature. In an effort to address this problem, FlyBase has introduced a new literature curation triage system that employs a mix of brief and in-depth examination of journal articles. During the triaging process, papers rich in functional data are specifically flagged for GO curation.
In addition to information about D. melanogaster, FlyBase includes genome sequence data, gene models and protein information for an additional 11 Drosophila species (3,4). While D. melanogaster remains the focus of our manual GO annotation effort, we also add GO terms for non-melanogaster species based on experimental evidence in the literature. The majority of manual GO annotations for non-melanogaster genes are provided by UniProtKB (5) curators, who include GO terms based on sequence similarity. In addition, we have recently expanded our automated GO annotation pipeline to include GO terms based on InterPro (6) domains for all 12 sequenced Drosophila species.
GO REFERENCE GENOME ANNOTATION PROJECT
The GO reference genome annotation project (http://www.geneontology.org/GO.refgenome.shtml) is a cross-database collaboration that aims to provide comprehensive high-quality GO annotation for every gene product in 12 diverse species—Arabidopsis thaliana, Caenorhabditis elegans, Danio rerio, Dictyostelium discoideum, D. melanogaster, Escherichia coli, Homo sapiens, Gallus gallus, Mus musculus, Rattus norvegicus, Saccharomyces cerevisiae and Schizosaccharomyces pombe (7).
For each gene product, the project aims to find all of the relevant GO terms based on the available primary experimental data and to remove those terms that have become incorporated into databases based on unsubstantiated statements.
In the early days of GO annotation, FlyBase and other GOC members captured ‘common knowledge’ from reviews and text books. While most of this information is correct, occasionally there are examples where the information cannot be traced to an experiment or the experiment is in a different species from that annotated and may not be true of the gene being annotated. GO reference genome curators have agreed, therefore, to a set of stringent annotation standards to avoid future GO annotation errors and to improve annotation consistency between groups. The resulting set of ‘gold standard’ annotations will be an improved resource for researchers in these species and can also be used to annotate other genomes.
In addition to applying the agreed standard to all new GO annotations, curators work together to annotate specific genes. This approach encourages discussion of annotation issues between curators and is efficient for developing new GO terms in a given area of biology. Each month curators from one of the participating databases choose 20 genes, the orthologous genes in the other species are identified and curators endeavour to assign all applicable GO terms based on published experimental data. When experimental data for all species are captured and all GO annotations conform to the new standards, the full annotation set is reviewed by a curator from one of the databases to check for potential errors, for example, does an outlying term in one species represent an interesting feature of biology or a curator error? Finally, gaps in knowledge for a species are filled based on sequence similarity to the experimentally characterized gene products, where applicable. It is generally safer to transfer molecular function and cellular component terms than biological processes (e.g. the term ‘flower development’ would not be added to a fly gene no matter how similar the gene products).
The current priorities for annotation are: homologs of human disease genes, genes that are highly conserved across species, genes involved in biochemical/signalling pathways, and topical genes shown to be of significant interest in recent publications. FlyBase has been contributing GO annotations to the project since it started in August 2006; over 200 Drosophila homologs of human genes have now been curated. Details of the genes examined to date and associated GO annotations in all species can be found on the GO website (http://www.geneontology.org/GO.refgenome.shtml); GO annotation for all species is searchable using AmiGO (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi).
GO ANNOTATION REFINEMENTS
Assignment of terms ‘inferred from sequence or structural similarity’ (ISS) is a potential source of errors within existing annotations. In a sequence analysis, for example, if the top hits are all kinases it may seem reasonable to assume that the query sequence is also a kinase. Chances are, however, that none of the matching sequences are experimentally characterized kinases, but simply look like other sequences called kinases and so on. With the expansion of genomic data, these inferences based upon inferences are becoming increasingly common and are potentially misleading. To eliminate errors from such transitive annotations, the reference genome group have agreed to limit GO annotation based on sequence similarity to experimentally characterized gene products. The gene product identified in supporting evidence (‘with’ column of the gene-association file) must itself be annotated with the same (or more specific) term assigned with an experimental evidence code. FlyBase has reviewed its current set of ISS annotations and found that, for annotations where the similar sequence is recorded, just over 100 were made to genes products that did not have a GO annotation based on experimental evidence codes (Table 1). These have all been revised to conform to the new annotation standards. The second part of the ongoing ISS review involves checking the terms for old annotations where the existence of a similar sequence was not recorded.
In the interests of focusing on experimental data and attributing terms directly to publications that contain that data, FlyBase no longer curates new GO annotations based on review articles. As GO annotations for each gene are revised, existing terms based on author statements are traced to the original publication (where possible). Occasionally no experimental support for the term can be found in Drosophila and the term is removed. Similarly, we no longer assign GO terms based on the names of gene products in records submitted to DNA or protein sequence databases. Finally, no new GO terms will be assigned to gene products based on meeting abstracts; this information is better captured from subsequent publications where the data are presented in full.
FlyBase supplements manual GO curation with electronically predicted terms. We have recently standardized our ‘inferred from electronic annotation’ (IEA) data such that it is based on a single source: a mapping between InterPro protein domains and GO terms (6). This manually generated mapping is under constant revision, partly based on feedback from GO curators, and is considered to be 91–100% accurate (8). GO annotations based on InterPro domains are now updated for every new release of FlyBase and the InterPro domain ID is now included in each annotation. IEA data from other sources, which were several years old and frequently redundant (e.g. terms based on Panther protein signatures which are now incorporated in InterPro), have now been removed. This has also eliminated potentially confusing discrepancies between the GO annotation sets available from FlyBase and the GOC. The GOC recommends that electronically predicted data be revised annually and, in an effort to enforce good practice, removes any annotations from submitted data sets with IEA evidence codes that are >1 year old.
Table 4 shows a summary of our current GO data for D. melanogaster. Although many of the changes in GO annotation policy are quite recent, we can already see an improvement. While absolute annotation numbers are relatively unchanged because of deleted IEA data, the number of annotations based on experimental evidence has improved dramatically compared to FlyBase release FB2006_01.
Table 4.
Comparison of total GO annotations in FlyBase releases FB2006_01 and FB2008_08 for all D. melanogaster genes (including those not yet located to the genome) by evidence type
| FB2006_01 | FB2008_08 | % change | |
|---|---|---|---|
| Experimental evidencea | 11 888 | 17 322 | +46% |
| Computational evidenceb | 14 946 | 15 487 | +4% |
| Author/curator statementsc | 16 711 | 16 506 | −1% |
| Electronic annotationd | 22 626 | 16 010 | −29% |
| No biological data availablee | 4997 | 5022 | +0.5% |
| Total annotations | 71 168 | 70 351 | −1% |
aAssigned with evidence codes: IDA, IPI, IMP, IGI, IEP.
bAssigned with evidence codes: ISS, ISA, ISM, ISO, RCA.
cAssigned with evidence codes: TAS, NAS, IC.
dAssigned with IEA evidence code.
eRoot GO terms assigned with ND evidence code (note that ‘ND’ is applied only to genes that have been assessed for functional data; it is not used for genes that have not yet been subject to GO curation).
USER FEEDBACK
Users are encouraged to give us feedback about GO annotation in FlyBase. In particular, do the terms assigned to your favourite gene represent an accurate summary of the literature? We welcome input from gene family experts to improve the coverage, consistency and accuracy of our annotations. Comments and questions about Drosophila GO data or any aspect of FlyBase can be made via our website (http://flybase.org/cgi-bin/mailto-fbhelp.html).
REFERENCING FLYBASE
We suggest FlyBase be referenced in publications by citing this publication and the FlyBase URL (http://flybase.org).
FUNDING
National Human Genome Research Institute at the National Institutes of Health (P41 HG00739); Medical Research Council (UK) (G0500293). Funding for open access charges: NIH NHGRI grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Eleanor Stanley at UniProtKB for contributing GO annotation to FlyBase, and Nick Brown, Lynn Crosby, William Gelbart, Thom Kaufman and Kathy Matthews for comments on the article. S.T. would like to thank the members of the Gene Ontology Consortium and particularly the GO Reference Genome Annotation Group for support with all aspects of GO annotation and ontology development.
REFERENCES
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson RJ, Goodman JL, Strelets VB, The FlyBase Consortium FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosophila 12 Genomes Consortium. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 4.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM, The FlyBase Consortium FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot: the manually annotated section of the UniProt KnowledgeBase. Methods Mol. Biol. 2007;406:89–112. [Google Scholar]
- 6.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Gene Ontology Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camon EB, Barrell DG, Dimmer EC, Lee V, Magrane M, Maslen J, Binns D, Apweiler R. An evaluation of GO annotation retrieval for BioCreAtIvE and GOA. BMC Bioinformatics. 2005;6(Suppl. 1):S17. doi: 10.1186/1471-2105-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]