Abstract
Dramatic increases in the throughput of nucleotide sequencing machines, and the promise of ever greater performance, have thrust bioinformatics into the era of petabyte-scale data sets. Sequence repositories, which provide the feed for these data sets into the worldwide computational infrastructure, are challenged by the impact of these data volumes. The European Nucleotide Archive (ENA; http://www.ebi.ac.uk/embl), comprising the EMBL Nucleotide Sequence Database and the Ensembl Trace Archive, has identified challenges in the storage, movement, analysis, interpretation and visualization of petabyte-scale data sets. We present here our new repository for next generation sequence data, a brief summary of contents of the ENA and provide details of major developments to submission pipelines, high-throughput rule-based validation infrastructure and data integration approaches.
INTRODUCTION
The race for ever higher-throughput nucleotide sequencing technologies places bioinformatics, and life sciences research in general, at the leading of edge of the development of infrastructure for the storage, movement, analysis, interpretation and visualization of petabyte-scale data sets. Nucleotide sequencing information from high-throughput machines is fed into the worldwide computational infrastructure through the archival services operated by the major bioinformatics service providers. This information forms the foundation for higher tiers of interpretation, such as assembly into complete genomes, gene structure annotation and mapping to known reference genomes and transcriptomes for quantitative expression analysis.
The European Nucleotide Archive (ENA), the collective name for nucleotide sequencing information archiving and presentation services at EMBL-EBI, comprising the EMBL Nucleotide Sequence Database and the Ensembl Trace Archive, lies therefore at the forefront of the European focus for petabyte-scale data strategies. In our newly established archive for next generation read data from the latest high-throughput machines, for example, the first 3 months of accepting submissions saw the receipt at EBI of 10 TB of data, representing an eighth the total volume of data accumulated in 28 years of activity so far. Predicted improvements to sequencing platforms such as the Genome Sequencer FLX from Roche/454, the ABI SOLiD platform and the Genome Analyzer from Illumina are poised to increase throughput many-fold further. Raw sequencing information flows forwards into various other components of ENA and into broader EBI services such as UniProt (1) and Ensembl (2). The challenges that developments such as these present to the ENA require us to consider new technical approaches to storage and retrieval of data (including bandwidth-beating solutions to overcome network constraints), significant developments to user tools to submit the data, sophisticated validation systems to overcome unmanageable demands on manual curation efforts and sophisticated tools to maximize data utility to users. In this article, we review briefly the status of contents of the ENA and focus on how we are rising to meet the challenges of petabyte-scale nucleotide sequence data over the coming years, through developments to data submission systems, rule-based validation systems, storage of raw next generation sequence data and integration with broader services at EBI as a means of organizing ENA data.
STRUCTURE OF THE ENA
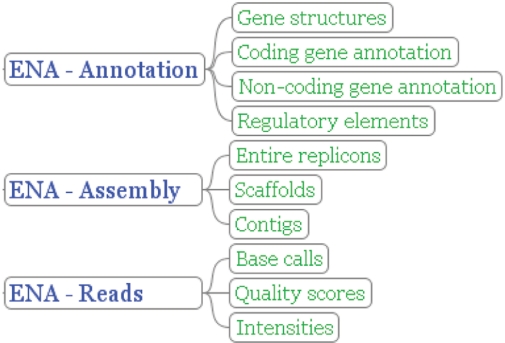
The breadth of information of interest to ENA is ever expanding both in terms of novel technologies for the resolution of nucleotide information and the applications to which the information is put. We have adjusted our overall view of nucleotide sequence archiving and have abstracted somewhat from underlying legacy infrastructure, such that sequencing information is classed as ‘reads’ (sequencing machine output—traces, flowgrams, etc.—base calls and quality scores), ‘assembly’ (information relating overlapping fragmented sequence reads to contigs and covering higher order structures where contigs are structured into representations of complete biological molecules, such as chromosomes) and ‘annotation’, where interpretations of biological function are projected onto coordinate-defined regions of assembled sequence in the form of annotation (Figure 1). In all cases, the core information is provided solely by submitters and is only updated by submitter interaction. This is in sharp contrast to the information in other databases, such as Ensembl, which provide a community view of the information provided. Associated with read, assembly and annotation information is information relating to the provenance and treatment of biological samples used for sequencing. In this scheme, the INSDC component contributes information to both ENA-Assembly and ENA-Reads. Where possible, data in ENA-Annotation, ENA-Assembly and ENA-Reads are connected in a single integrated system, such that links can be made between data objects in each of the components (e.g. annotation on highly assembled sequenced can be tracked back to underlying contigs and capillary trace data that support a particular assembly can be retrieved).
Figure 1.
ENA structure. The figure shows how nucleotide sequencing information is partitioned according to class; ENA-Reads treats raw sequencing information, ENA-Assembly treats information on how fragmented sequences have been assembled into higher order structures and ENA-Annotation treats functional annotation based on assembled sequence. The three components are integrated in the ENA.
As an archival repository, the primary information stored and presented is derived from the submitting parties; ownership, and hence editorial control, of primary content, remains with the original submitting group. However, an archive of such size and diversity clearly requires sensible organization of data for management purposes and end-user utility (such as search and visualization) and integration with the many other tools and data resources available at EBI and beyond. Such data organization and integration require active mapping maintenance between ENA objects and objects in remote resources. Developments in these integration pipelines are discussed subsequently.
The ENA achieves comprehensive coverage of the world's nucleotide sequencing data through a number of active collaborations, most notably with DDBJ (3) and GenBank (4), though the INSDC (The International Nucleotide Sequence Database Collaboration, http://www.insdc.org) and through trace collaborations with the Wellcome Trust Sanger Institute and the trace archive at NCBI (5). As part of our drive to improve the utility of archived data, we are active in the development of a number of formats and standards, including MIGS (6), CBoL BARCODE data standard (http://barcoding.si.edu/PDF/DWG_data_standards-Final.pdf) and MINSEQE (http://www.mged.org/minseqe/MINSEQE.pdf).
ENA provides comprehensive submission tools and services, permanent archiving of content and a multitude of data access resources. Points of entry into ENA services are detailed in Table 1.
Table 1.
Points of entry to the ENA: submissions, retrieval and support
| Submissions | |
| Submission of new data http://www.ebi.ac.uk/embl/Submission/webin.html | Submissions of annotated sequence data to ENA-Annotation and capillary traces to ENA-Reads |
| Updates to existing data http://www.ebi.ac.uk/webin/update.html | Updates to existing ENA-Annotation data |
| Next generation sequence, project accounts and WGS submissions datasubs@ebi.ac.uk | To establish next generation sequence submissions, new project accounts and pipelines for WGS projects. |
| Retrieval | |
| SRS http://srs.ebi.ac.uk | Data retrieval by term search and through links to/from other databases |
| Sequence similarity search http://www.ebi.ac.uk/Tools/similarity.html | Data retrieval by sequence similarity |
| Sequence Version Archive http://www.ebi.ac.uk/cgi-bin/sva/sva.pl | Access to current and previous versions of entries by accession number |
| ENA-Annotation and ENA-Assembly FTP ftp://ftp.ebi.ac.uk/pub/databases/embl/ | Access to complete data sets in flatfile format, for both release and updated data |
| ENA-Reads FTP ftp://ftp.ensembl.org/pub/traces/, ftp://ftp.era.ebi.ac.uk/ | Access to ENA-Read data for capillary and next generation reads, respectively |
| Genomes http://www.ebi.ac.uk/genomes/ | Completed genomes and proteomes |
| Dbfetch/Wsdbfetch http://www.ebi.ac.uk/cgi-bin/emblfetch, http://www.ebi.ac.uk/Tools/webservices/WSDbfetch.html | Retrieval by accession number through web browser, or via webservice, respectively |
| Custom Datasets datasubs@ebi.ac.uk | For data retrieval requirements not supported by existing tools |
| Support | |
| General Information http://www.ebi.ac.uk/embl/ | Documentation including user manual, INSDC Feature Table Definition, news and forthcoming changes |
| Specific Help datasubs@ebi.ac.uk | For help with all ENA services |
| Educational Information http://www.ebi.ac.uk/2can/ | Background bioinformatics educational resources |
CONTENTS IN 2008
At the time of writing, ENA contains 143 million ENA-Annotation records, covering 233 billion bases and ENA-Reads contains 1.8 billion bases of capillary reads with 10 TB of next generation sequence data. In all, 400 000 different taxonomic nodes are connected to sequence, over 200 000 published papers are explicitly cross-referenced in ENA records and ENA maintains 99 million cross-references to objects in external resources.
Notable high-volume data sets new in 2008 include the raw sequencing data (460 Gbases) from the genomes of 83 individuals from the ongoing human 1000 Genomes Project (ERA000013-ERA000026), all of the underlying data for an extensive genome variation and evolution study including 17 Salmonella typhi isolates [WGS accessions starting CAAQ-CAAZ and ENA-Reads accession ERA000001, ref. (7)] and two newly sequenced genomes from a trio of isolates, including an important drug-resistant nosocomial isolate of Acinetobacter baumannii [ENA projects 13001 and 28921, ref. (8)].
SUBMISSION SYSTEMS
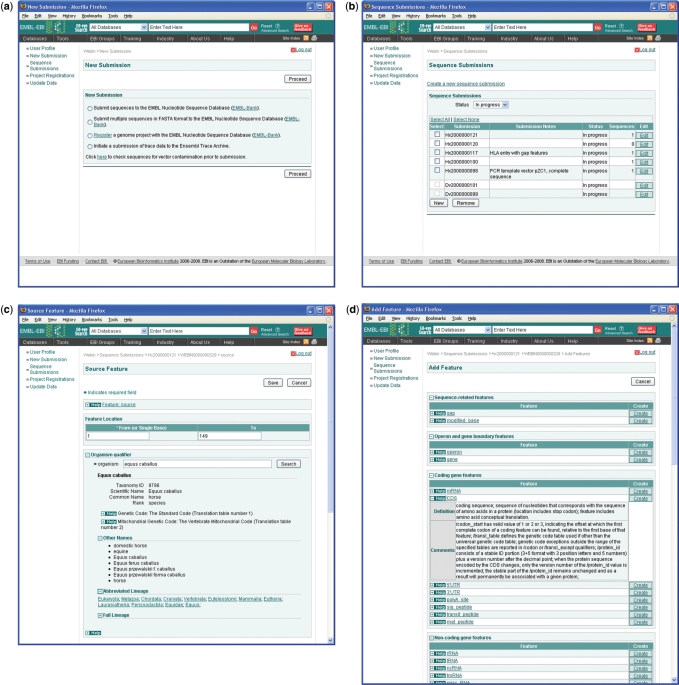
Late in 2008, we launched significant improvements in functionality in our submission systems, both for the submission of small sets of annotated sequences, in a public beta testing service, and for the submission of metadata describing next generation sequencing experiments, in a production service. The first has been made possible by an ongoing replacement of our underlying core submissions infrastructure that was initiated some time ago with aim to provide such facilities as robust rule-based validation (see subsequently), extensibility and support for large-scale genome submissions. Our future plans include the migration of the remaining submission types across to the new infrastructure and significant improvements to functionality. We expect that these improvements will provide us with the capacity to continue to maintain and improve upon our traditional quality standards. A single entry point into the submissions system (Figure 2a) provides triage of submissions based on a limited number of straightforward user choices and despatches users to the appropriate service.
Figure 2.
Webin. The figure shows a selection of screenshots from Webin; (a) launcher page, (b) submissions page, (c) source feature page and (d) new feature addition page.
Small-scale submissions through Webin
Interactive web applications are well established as the methods of choice in the submission of small-scale data sets, particularly from infrequent and non-expert submitters who have limited knowledge of ENA data structures and limited bioinformatics expertise. For 10 years, the perl-CGI Webin application has provided the submission tool for over a million sequence-annotation entries. While updates to ENA flatfile structure and to the INSDC Feature Table Definitions have been progressively implemented and maintained, the system lacks the flexibility for rapid additions to functionality that are available with framework web technologies available today.
Available in the 2008 beta release of Webin are an integrated Lucene-based taxonomy browser and search facility (Figure 2c), capable of resolving taxonomic names and their synonyms and of providing visualization of taxonomic classifications; an integrated Feature Table Definition browser (Figure 2d), that allows users context-dependent reporting of the latest definitions, value formats, usage examples and comments for features and qualifiers; improved grouping of features and qualifiers in annotation pages (Figure 2d), that abstracts submitters from the syntax of the Feature Table Definitions, thus guiding submitters through their submissions, and a rule-based validation system.
Next generation data submissions
We have established two pipelines for the submission of sequence data from next generation sequencing machines. The highly normalized structure of this part of ENA (see subsequently) allows for completely separate treatment of the large data components and the much smaller metadata components. Data files, prepared in Sequence Read Format (http://srf.sourceforge.net) or native machine formats, are submitted with limited need for input from the submitter and metadata are submitted with greater interaction between the EBI and the submitter, reflecting the high degree of validation that can be achieved.
Small-scale submitters currently alert the ENA team by email (Table 1) that they have a submission pending and a private FTP drop box is created for them. A notification email is sent to the submitter providing instructions and details of the drop box. At this stage, the drop box contains a pro forma spreadsheet ready for completion with details of samples, experimental procedures and run configurations. Where available, any information gathered from the initial contact with the submitter is already completed in the spreadsheet. Included in the information collected at this point are MD5 values that the submitter has calculated for the data files to be submitted. ENA re-calculates these checksums as a cross-check when the data files are submitted. When submitters have completed the spreadsheet, they upload with their data files to their FTP drop box. In many cases, submitter-limited bandwidth restricts this data transfer using FTP, so ENA is able to accept hard disk submissions by return-paid courier services. Once files have been received, the ENA team validates, accessions and uploads the data into the production database. In due course, we will integrate the spreadsheet functionality into our interactive Webin submission tool.
Large-scale submitters, mainly from the sequencing centres, typically run LIMS systems, in which many pieces of information that are ultimately required as metadata for a submission to ENA have already been stored for tracking purposes. This is particularly true for study-, sample- and run-level information. For these submissions, we receive metadata in XML format (see Table 1 for links to documentation) through a webservice that was designed in collaboration with the European sequencing centres. The webservice provides a management tool for the upload and tracking of both XML metadata files and data files. In order to maximize the network bandwidth available for transfer of data files, Aspera software (http://www.asperasoft.com/technology/index.html) is used to enhance network throughput.
RULE-BASED VALIDATION REPAIR TO IMPROVE THROUGHPUT
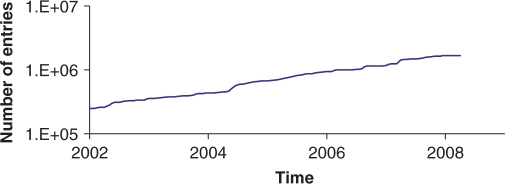
The ENA has long recognized the growing demand on its biological curation staff placed by increasing data volumes; indeed increasing volumes put pressure on all workflows that require manual intervention. This pressure is most notable in data input workflows for ENA-Annotation data, where biological curation is applied to incoming data to ensure consistency and optimal downstream utility. While our strategy for continual review and automation of workflows when technology and knowledge becomes available has thus far enabled us to cope with volume growths (Figure 3, where it can be seen that the throughput of validated ENA-Annotation records is seen to increase steadily), we plan to continue to apply this strategy and to focus strongly on the requirement to capture only primary information that is useful and cannot be calculated from ENA data (9). A guiding design principle that has already been useful has been that biological knowledge is most efficiently applied in ways that impact upon multiple records. In highly repetitive data sets, such as those from population biology studies, ENA biologists currently carry out limited editing of individual database records, but instead edit ‘submission master’ records containing all of the invariant information; information that varies between records is then later ‘inserted’ into master records to generate independent database records for each submitted sequence. Using this workflow, a single application of biological knowledge impacts upon multiple records within a submission. A future extension of this, currently under development, is a rule system in which the biologist curates a set of rules, each of which can be applied to a class of data. An example might be a rule constraining the use of a particular annotation structure to a given evolutionary clade. With such a rule system, a single application of biological knowledge impacts upon multiple records from multiple submissions (see Table 2 for a selection of sample validation rules).
Figure 3.
Throughput for validated ENA-Annotation entries. The figure shows cumulative counts of ENA-Annotation entries that have been processed by ENA biologists.
Table 2.
Sample validation rules
| Condition 1 | Condition 2 |
|---|---|
| QE(/environmental_sample) | QE(/isolation_source) |
| QE(map) | QE(/chromosome) OR QE(/segment) OR QE(/organelle) |
| QE(/proviral) OR QE(/virion) | NOT (QE(/proviral) AND QE(/virion)) |
| ME(‘BARCODE’) | QE(/pcr_primers) |
| QC(/organisms, ‘Bacteria’) AND NOT QE(/environmental_sample) | QE(/strain) |
| NOT (QC(/organism, ‘Deltavirus’) OR QC(/organism, ‘Retro-transcribing viruses’) OR QC(/organism, ‘ssRNA viruses’) OR QC(/organism, ‘dsRNA viruses’)) | NOT QV(/mol_type, ‘genomic RNA’) |
Sample validation rules are shown; a grammar has been developed that allows non-technical curation of combinations of conditions that should be satisfied in combination in order for the rule to passed. The QE function expresses the existence of a source qualifier of the specified name, the ME function expresses the existence of the specified methodological keyword, the QC function expresses parity or a child relationship to the specified value within the taxonomic hierarchy and the QV function expresses a specified value for the specified source qualifier.
The first deployment of the rule system in the Webin submission application takes advantage of the system's error-reporting component, that provides structured error information (e.g. which rule has been broken and in which way), and its repair component, which executes rule-specific repair actions, where these are possible, upon the data such that the rule is subsequently satisfied. Error reports and repairs are structured such that they can be interpreted differently according to the application that has called them; in the case of Webin, for example, errors can be reported to the user through text boxes that appear alongside submitter-controlled fields that need to be addressed in order to satisfy the rule or repairs can be executed with the appropriate warning text (that a change has been made to a field), in cases where sufficient information exists to make the appropriate repair.
Establishing a rule-based system for validation repair has proven useful already in the context of rapid and uncomplicated update of validation procedures within the Webin submission system detailed above. However, the utility of the system has far broader future applications. For data input pipelines from the large sequencing centres, in one example, ENA has traditionally relied upon the submitter's local bioinformatics resources to generate database-ready files for loading with limited input from EBI biologists. In a second example, existing legacy data may have content that has been brought into question following some new biological finding. A deployment of a validation repair tool in these scenarios, then, will yield significant data quality improvements.
ARCHIVING NEXT GENERATION READ DATA
Next generation sequencing technologies yield unprecedented data volumes through a combination of high-throughput microfluidics and novel sequencing chemistry. They are characterized by very highly parallel anonymous treatment of pooled samples, comparatively short reads (currently ∼35 for Illumina and SOLiD, ∼100 for 454, compared to ∼900 for ABI capillary) and low per-read cost. These characteristics have opened nucleotide sequencing to a broad range of applications, going far beyond the conventional use of sequencing in the determination of genome and transcriptome sequences for the purposes of assembly and subsequent functional annotation, that include expression analysis, re-sequencing for polymorphism discovery, epigenomics and gene discovery. Due to this diversification of applications, the ENA-Reads archive for raw sequencing data will provide not only access to data for the ENA-Assembly and ENA-Annotation framework, but will also serve as a public- and service-facing resource in its own right, through text search of metadata, a high-volume sequence similarity search and an API to access the data themselves. Internal users will include the Ensembl Functional Annotation project (2) and ArrayExpress (10).
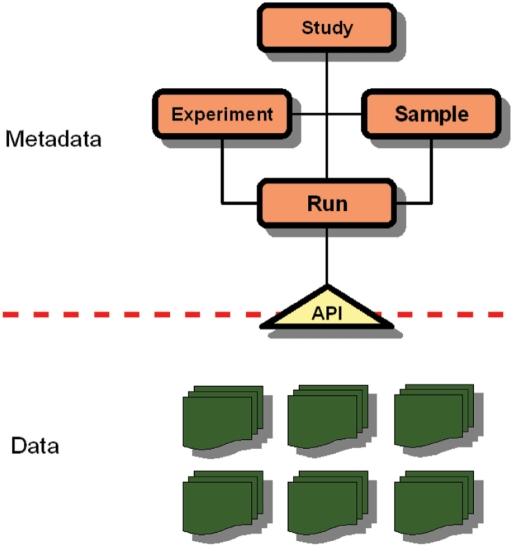
The parallel nature of the output of next generation sequencing machines has led to a radical re-think of the model for storing raw sequence data that has been adopted for conventional capillary reads. While it has been possible to store these latter in direct association with their metadata (sample preparation information, taxonomic information, library name, etc.), such a flat structure cannot scale appropriately. We have designed a hybrid file-database system (Figure 4) that maximizes the normalization of the information and allows us to take advantage of the efficiencies of file systems for large data volumes and the flexibility of a relational database for metadata access and manipulation. The data file component of the system takes advantage of a high degree of compression and non-tape backup systems to provide long-term security. The metadata component takes a more conventional relational schema, comprising individually accessioned study, experiment, sample and run objects to allow for flexible querying and metadata browsing. The accessioning system allows the reuse of existing objects, such as the reuse of a sample object in a new sequencing experiment. The two components communicate through an API that provides a layer of abstraction between the data files themselves and the user (internal or external). The API provides access to specified selections of intensity, sequence and quality data for entire machine runs, for sets of reads and for individual reads, abstracted from any grouping that may have been applied when data were generated and submitted. Reads are accessioned individually and, in combination with submitting centre name, are unique across the archive.
Figure 4.
Structure of ENA-Reads. A relational data model has been developed for next generation sequencing data that relates the concept of a study to samples that have been used for the study, to runs that have been executed as part of the experiments that make up the study and describe the details of how samples have been configured in runs. Underlying this data model is an API that provides abstraction from the nature of the data file system, returning read data upon request based on read identifiers (and groupings of these identifiers), rather than on specified files within the file system.
ENA-Reads is populated through submissions (see above) and through data exchange with the Short Read Archive at the NCBI (5). Data exchange is made possible by the adoption of similar infrastructures at each institute. Data file exchange between institutes is continuous to maximize available bandwidth across networks and metadata exchange takes place on a daily basis. Data are accessioned within the same namespace at either institute such that the same identifiers are used regardless of where a user goes to retrieve data.
At the time of writing, all ENA-Reads data can be accessed through the FTP site (ftp://ftp.era.ebi.ac.uk/), classified by submission, with metadata and data presented in XML and SRF or native formats, respectively. We expect soon to be able to replace this service with a more flexible metadata browser and search tool in combination with public access to the data file API.
INTEGRATED DATA PRESENTATION
Amongst the challenges of the high volumes of nucleotide sequence data available is the presentation of those data in useful ways to users. Central to our approach to this is the integration of ENA data with other resources, both those that provide information relating to the source and preparation of sequencing data and those that take ENA data as source and provide analysis and interpretation. Conventional cross-references in ENA provide channels through which users can get to appropriate ENA data (e.g. return of primary nucleotide sequence data starting from a transcript record in Ensembl) and it provides those who have resolved a given ENA record or set of records links away from ENA into resources that have additional information on the records (e.g. return of protein functional information in UniProt starting from a CDS feature in ENA-Annotation). However, in a further approach, integration also allows us to organize our data into sensible and useful portions by maintaining a web of overlapping groupings of objects in ENA; this represents a projection of analysis and interpretation information from the secondary resource back onto ENA records.
A first example of this use of integration exploits the paradigm that the genome sequence of an organism provides the most sensible structure around which to organize biomolecular data; since most ENA data are associated with organisms that have, or will soon have, completely sequenced genomes and since integration with non-nucleotide data provided by EBI also takes this paradigm, this is a sensible approach. In 2008, we introduced grouping of ENA-Annotation records by gene and by transcript for the major Ensembl species, which impacts upon in the order of 400 000 ENA records. Conventional cross-references from ENA-Annotation transcript records to Ensembl genes and transcripts have been implemented in cases where the ENA transcript has been used as supporting evidence for a computationally predicted gene in Ensembl; these cross-references use the existing ‘DR’ (‘Database cross-Reference’) line shown in ENA-Annotation flatfiles. Further cross-references, however, have been introduced that import the Hugo Gene Nomenclature Committee [HGNC, ref. (11)] or Mouse Genome Informatics [MGI, ref. (12)] gene symbol assigned to the Ensembl gene into the ENA record. While this offers additional non-primary information that may be of use while viewing ENA records, more importantly, it allows search tools built upon the records to index by not just a submitted gene symbol, but by an actively maintained and tracked community consensus gene symbol. The SRS tool at EBI, offers search of ENA transcript records by Ensembl-tracked gene symbols.
Further examples of this type of organization exploit existing integration with literature resources (through ‘RX’ lines), taxonomy (through ‘/organism’ values and taxonomic divisions) and INSDC project records (through ‘PR’ lines) and possible future integration with strain identifiers (through ‘/culture_collection’ values with the StrainInfo service, http://www.straininfo.net), non-coding RNA classifications (through ‘/ncRNA_class’ values with RFAM, http://www.sanger.ac.uk/Software/Rfam/) and many others. Ultimately, we intend to offer browser functionality that will allow users to browse directly across to peer ENA records (for example, to other ENA records that have also been used as supporting evidence for Ensembl genes and transcripts or to other records from the same strain). Ultimately, we intend to extend this functionality to handle intersections between overlapping groupings (for example, to allow a user to browse from one ENA record across to other ENA records that provide supporting evidence and are from the same strain).
FUNDING
European Molecular Biology Laboratory and the Wellcome Trust. Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
REFERENCES
- 1.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugawara H, Ogasawara O, Okubo K, Gojobori T, Tateno Y. DDBJ with new system and face. Nucleic Acids Res. 2008;36:D22–D24. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, et al. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS ONE. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochrane G, Akhtar R, Aldebert P, Althorpe N, Baldwin A, Bates K, Bhattacharyya S, Bonfield J, Bower L, Browne P, et al. Priorities for nucleotide trace, sequence and annotation data capture at the Ensembl Trace Archive and the EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2008;36:D5–D12. doi: 10.1093/nar/gkm1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M, et al. ArrayExpress-a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA the members of the Mouse Genome Database Group. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]