Abstract
Gene transcription is largely regulated by sequence-specific transcription factors (TFs). The TF activity is significantly regulated by its posttranslational modifications (PTMs). TF-PTMs serve as ‘molecular switchboards’ that map multiple upstream signaling events, in response to various environmental perturbations, to the downstream transcriptional events. While many instances of TF-PTMs and their effect on gene regulation have been experimentally determined, a systematic meta-analysis or a quantitative model-based investigation of this process has not been undertaken. A prerequisite to such analyses is a database of known instances of TF-PTMs affecting transcriptional regulation. The PTM-Switchboard database meets this need by cataloging such instances in the model system Saccharomyces cerevisiae. The database stores triplets of genes such that the ability of one gene (TF) to regulate a target gene is dependent on one or more PTMs catalyzed by a third gene (modifying enzyme). The database currently includes a large sample of experimentally characterized instances curated from the literature. In addition to providing a framework for searching and analyzing the data, the database will serve to benchmark computational methods. In the future, the database will be expanded to mammalian organisms, and will also include triplets predicted from computational approaches. The database can be accessed at http://cagr.pcbi.upenn.edu/PTMswitchboard.
INTRODUCTION
Gene transcription is regulated, in large part, by transcription factor (TF) proteins that bind to the relative vicinity of the target gene in a sequence-specific fashion. The activities of the TFs themselves are often regulated through one or more posttranslational modifications (PTMs). PTMs constitute covalent chemical changes—e.g. phosphorylation, acetylation, methylation or glycosylation—to the residues in the protein sequence. Hundreds of distinct types of PTMs have been reported (1,2), at least a dozen of which regulate TF activities (3–6).
TF-PTMs modulate, through mechanisms that are not completely known, the activity of TFs and the downstream regulation of the target genes, thus acting as ‘molecular switchboards’ that map inputs from cell signaling pathways to gene transcripts (3,4). On one extreme, such regulation can be simple and binary—i.e. PTMs that serve as ‘on/off’ switches for TFs. More often, however, this regulation is highly complex, with multiple signaling inputs integrated into tightly regulated transcript levels. Furthermore, the relative concentrations of many PTMs at steady state are dynamically maintained by opposing forces catalyzing the addition or removal of a chemical group. To fully understand the complexity of TF-PTM-mediated regulation of gene transcription, we first need to map the connectivity between signaling molecules, PTMs, TFs and target genes, and such a map currently does not exist. The characterized instances of TF-PTMs are biased largely towards well-characterized TFs, e.g. the cAMP-response element binding protein (CREB) (7), and PTM types, e.g. phosphorylation (8). Therefore, we cannot begin to explore the more complex problems of molecular kinetics or network topologies surrounding TF-PTMs on a systematic basis.
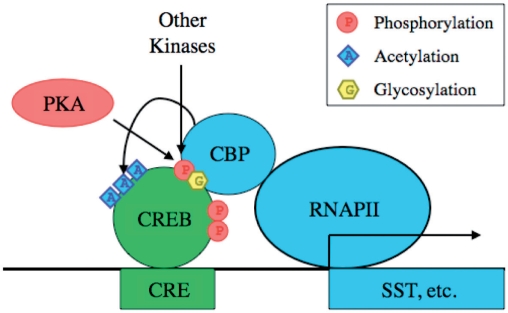
A canonical example of a regulatory TF-PTM is Ser133 phosphorylation of CREB in mammals (Figure 1). This TF-PTM has long been characterized as a key event in protein kinase A (PKA) signaling that results in the activation of target genes. The primary serine phosphorylation allows CREB to interact with its co-activator, CBP, thereby recruiting the core transcriptional machinery. Other kinases have since been discovered to also activate downstream genes through this same TF-PTM. Other PTMs that alter CREB activity have also been discovered, including additional phosphorylations, acetylations and interestingly, an O-linked N-acetyl glycosylation (O-GlcNAc) that antagonizes the primary activating phosphorylation. These modifications are reviewed in further detail in ref. (3). CREB exemplifies the potential complexity of TF-PTM regulatory programs beyond simple ‘on/off’ switches, and the emerging appreciation for modifications other than phosphorylation.
Figure 1.
cAMP-response element binding protein (CREB) Regulation. CREB is a well-studied TF that exemplifies the complexity of TF-PTM regulatory circuits. Canonical CREB regulation begins with phosphorylation of Ser133 by Protein Kinase A (PKA), which facilitates interaction with CREB-binding protein (CBP) to recruit RNA Polymerase II (RNAPII) and promote transcription of target genes such as Somatostatin (SST). Other kinases can also regulate CREB through Ser133 and other phosphorylations, CBP can further regulate CREB activity through multiple acetylations, and glycosylation can disrupt the activating effect of the Ser133 phosphorylation.
Few TFs have been studied as extensively as CREB, and numerous TF-PTMs remain to be discovered. To investigate TF-PTM-mediated gene regulation at the level of systems biology, we first need to identify the key regulatory connectivity between modifying enzymes, TFs and target genes, as well as the specific TF-PTMs involved in transducing these signals. The potential combinations of modifying enzymes, TFs, and target genes, even in simple organisms such as S. cerevisiae, are overwhelming. However, only a miniscule portion of these combinations are relevant to gene regulation, and prioritizing them for further study is a key step towards modeling this complex form of regulation.
A comprehensive view of the TF-PTM landscape could provide clues to broader biological questions, for instance: (i) the process-specific roles of individual types of PTMs, especially the types that are not as well studied as phosphorylation, such as hydroxylation (9), and (ii) the common mechanisms and principles by which TF-PTM circuits function and evolve. Our ability to approach these questions is limited by our knowledge of experimentally determined TF-PTMs and their effect on the transcription of individual genes. Predictive approaches have been proposed to identify cases where the regulatory interaction between a TF and a target gene is dependent on the expression of a modifier gene (10–12). Such genome-scale predictions could be used to construct a ‘rough map’ of the TF-PTM landscape to begin to answer the questions above. However, these predictive methods are in their infancy and lack a ‘gold set’ by which to gauge their performance. As one of its goals, PTM-Switchboard will provide a benchmark set for predictive methods.
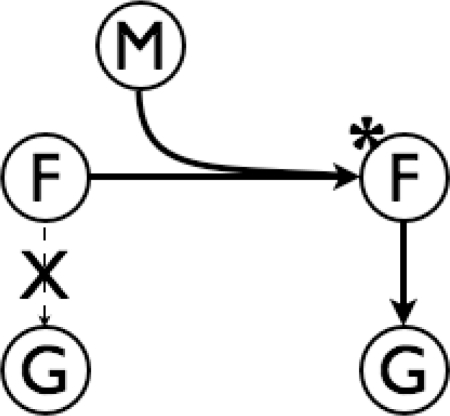
PTM-Switchboard differs from existing molecular pathway databases (13–15). Instead of using pair-wise interactions as a primary data type, PTM-Switchboard stores triplets of genes such that the ability of one gene (the TF) to regulate a target gene is dependent on a third gene (the modifying enzyme). We refer to this as the Modifier-Transcription Factor-Gene triplet, or in short, the ‘MFG triplet’ (Figure 2). The database currently contains 116 experimentally validated instances covering 13 modifying enzymes, 11 TFs and 91 target genes—a sufficient set to evaluate computational approaches and seed further cataloging efforts.
Figure 2.
Example MFG Triplet. In this simple example, a factor (F) is unable to regulate the target gene (G) in its initial state. Modification catalyzed by the enzyme (M) transitions F to a new state (marked by *) at which point it is able to regulate G.
PTM-Switchboard is also intended to seed a larger community effort to build a more comprehensive database of MFG triplets, as they are extremely laborious to search and curate from the literature. Text-mining approaches (16,17) are currently limited to identifying pair-wise interactions, and MFG triplets are rarely studied together as part of a single paper. More commonly, the overall effects of a PTM on a TF's cellular localization, degradation or DNA-binding activity are studied in one reference, while the gene targets of the TF are studied independent of any PTMs in another reference. In some cases, MFG triplets can be inferred from these references together, but only with careful consideration of the molecular mechanisms involved—a task clearly beyond current text-mining methods. PTM-Switchboard links the two fields of cell signaling and transcriptional regulation, and provides substantial opportunity to leverage the existing knowledge bases in these fields.
DATABASE DESIGN AND CONTENT
The primary unit of data used in PTM-Switchboard is a three-way interaction—the MFG triplet—as described above and illustrated in Figure 2. The specific guidelines for including MFG triplets in this database are further clarified below. Using the MFG triplet representation, the effect of a modifying enzyme can be defined for each of a TF's target genes individually, rather than uniformly across all TF targets. Triplets contained in the database may share one or two members. For example, Hog1 regulates the overall transcriptional activity of Sko1 at a set of target gene promoters, and separate triplets are included for each target.
In general, TF activity at a gene promoter can be modulated by a variety of proteins, including signaling molecules, co-factors and chromatin-modifying enzymes. A ‘Regulatory Triplet’ constitutes a TF, its target gene and a ‘modulator’ gene, which alters the TF's target-specific activity. MFG triplets differ from other regulatory triplets primarily in the physical interaction between the modifying enzyme and the TF, which yields a covalent chemical change in the TF protein. This has several important implications for specifically studying MFG triplets, including: (i) the possibility that the modifying enzyme regulates transcription distally, i.e. prior to the TF–DNA interaction and outside the nucleus; (ii) the implication that transcript expression data alone may be insufficient for studying MFG triplets, due to posttranscriptional regulation of both the TF and the modifying enzyme; and (iii) a unique set of additional data involving the detection and modeling of enzyme–substrate interactions and PTMs (18,19) can be applied to the study of MFG triplets.
PTM-Switchboard specifically focuses on MFG triplets involving a direct catalysis of a TF-PTM by a modifying enzyme, and does not include other types of regulatory triplets or the cases that lack evidence for direct catalysis. Furthermore, the PTM must be relevant to TF activity at the promoter of the included target gene. This basic unit is an essential building block for more advanced studies of kinetics and fine-scale regulation at both the transcriptional and posttranscriptional level.
Each PTM is recorded at a specific residue of a TF, but independent of the modifying enzyme responsible. The same PTM can be involved in multiple MFG triplets, and likewise an MFG triplet can involve multiple PTMs. MFG triplets can involve both addition and removal of a chemical group. Even in the well-studied cases, the exact positions of the PTMs often have not been mapped. However, PTM-Switchboard provides a framework to store these PTMs as they are identified or hypothesized.
PTM-Switchboard includes additional biological information about the MFG triplet. The modifier can either have a positive or negative effect on the activity of the TF, and likewise the TF can be an activator or repressor of each target gene. In some especially complex cases, such as Sko1, the TF can act as both an activator and a repressor, depending on the activity of the modifying enzyme—in this case, kinase Hog1 (20). To describe the behavior in such cases, the overall activity of the triplet is described by the influence of the TF on the target gene (positive, negative or neutral) in both the cases when the modifying enzyme is active and when the modifying enzyme is inactive. For example, in Figure 2 the relationship between F and G is neutral when M is inactive, and positive when M is active. Note that this is still a simplification of complex dynamics, and is intended to serve as a primer for more quantitative investigations.
PTM-Switchboard is also an exploratory tool for molecular biologists, and provides a considerable amount of supporting data for the curated instances of the MFG triplets, including links to external annotation resources (14,21). All genes are recorded using both their gene symbol and ORF ID according to the Saccharomyces Genome Database (SGD) (22), thus directly linking each gene to its SGD annotation page. All information contained in PTM-Switchboard is derived either from literature or other knowledge bases, and the sources used to derive this information are continually tracked and annotated, as discussed in the next section. A detailed description of the curation process is provided in the Supplementary Material.
SUPPORTING EVIDENCE
The MFG triplets currently contained in PTM-Switchboard vary in their mechanistic complexity and are supported by heterogeneous experimental evidence. To help the user assess each triplet, the database includes an extensive annotation of the sources for each MFG triplet. The most convincing cases are those in which all three genes are studied together—i.e. the effect of perturbing the modifying enzyme is studied on both the TF and the target gene—using a combination of genetic and biochemical techniques.
While MFG triplets identified through focused biochemical experiments garner the highest confidence in their accuracy, there is also a great deal of knowledge to be gained from high-throughput and genetic experiments, even if these experiments occasionally provide invalid MFG triplets. Genetic experiments alone can detect MFG triplets, but cannot distinguish them from other forms of regulation and network connectivity. For example, the modifying enzyme may be further upstream from the TF in the signaling pathway, or likewise the known TF may target another TF, which then targets the gene of interest. Many kinases are also known to operate as co-factors, i.e. by binding TFs at promoters to help recruit, activate, or block the core transcriptional machinery (23–25). Such mechanisms do not qualify for inclusion in PTM-Switchboard, however any triplet is useful to include in the database if the best model explaining the genetic evidence involves a TF-PTM. Therefore, PTM-Switchboard has been designed to integrate current knowledge from all available sources.
To address the potentially subjective nature of weighing different evidence types, we have developed a system of tagging each triplet with a set of codes that briefly and concisely describe the evidence contained in the literature. This allows the user to quickly assess their confidence in individual triplets based on their own criteria without having to examine each literature reference. In the future, we will add triplets predicted by text-mining and computational modeling approaches, which can be experimentally validated. Appropriate evidence codes can be assigned to separate the “gold set” from computational predictions. A complete set of the evidence codes currently used is shown in Table 1.
Table 1.
Evidence codes for the MFG triplet in the database
| Code | Definition |
|---|---|
| V | Experimentally demonstrated in vivo |
| T | Experimentally demonstrated in vitro |
| L | Identified in low throughput experiment |
| H | Identified in high throughput experiment |
| P | Identified in computational prediction |
| C | Supported by biochemical experiment |
| G | Supported by genetic experiment |
| I | Inferred from multiple sources |
| S | Demonstrated in a single experiment |
| M | Identified from literature by text-mining |
| H | Identified from literature by human curation |
Of particular interest is the ‘I’ code, which indicates that the triplet was inferred from a combination of literature references. For instance, one paper may demonstrate that a modifying enzyme regulates a TF's overall activity under a specific condition, i.e. by changing its nuclear import/export, while a second paper may demonstrate that the TF is known to regulate a target gene under a similar condition. Triplets marked with the ‘I’ code indicate that the experimental conditions in the referenced papers were comparable enough to justify combining their results. The ability to integrate multiple references is a major advantage of expert curation.
In addition to the gold set of 116 fully characterized and experimentally validated genes, we are also using PTM-Switchboard to track incomplete MFG triplets where computational predictions may be of the most immediate value to molecular biologists. These typically occur in the literature when perturbation of a modifying enzyme leads to a significant change in another gene's transcript level, but the TF transducing the signal is unknown. Likewise, a PTM may be known to influence the activity of a TF, but the enzyme catalyzing the addition or removal of this PTM may be unknown. These entries are marked with the evidence code ‘U’ to mark them as having exactly one ‘unknown’ member and they are excluded from the ‘gold set’ for validating prediction algorithms.
CASE STUDY: THE FUS3/STE12/FUS1 TRIPLET
Here, we present one example of PTM-mediated transcriptional regulation included in PTM-Switchboard that highlights the mechanistic complexity of this process and thus the value of expert curation. Additionally, this case study illustrates the guidelines for inclusion of a MFG triplet in the database and the benefit of using evidence codes. Activation of the mitogen-activated protein kinase (MAPK) Fus3 is known to up-regulate the transcription of many genes, including FUS1, via activation of the Ste12 TF at pheromone response elements in gene promoters (26). Yeast mating factors trigger this response via a well-studied signaling pathway culminating in this MFG triplet (27–29).
On the surface this appears to be a straightforward MFG triplet, with the simplest model suggesting that Fus3 activates Ste12 via phosphorylation. However, a closer inspection of the literature reveals alternative regulatory mechanisms, including inhibitory phosphorylation of the Ste12 repressors Dig1 and Dig2 (28) and direct repression of Ste12 protein by inactive Fus3, which is lifted upon Fus3 activation (25,30). Most experiments focused on this triplet are genetic in nature, and cannot distinguish between these mechanisms. A small fraction of the papers contain biochemical evidence demonstrating that active Fus3 also phosphorylates Ste12 (31) in a way that promotes target gene expression (32,33). Therefore, this MFG triplet was included in the database, but without such evidence, the triplet would not have been included. Furthermore, triplets involving Dig1 and Dig2 as ‘modulators’ of Ste12 do not meet inclusion guidelines for the database regardless of evidence. In a similar case, there is genetic evidence for activation of Ste12 by another MAPK, Kss1 and the most widely accepted model for this evidence includes phosphorylation of Ste12 by Kss1 (25). In this case, a set of KSS1/STE12 triplets is included in PTM-Switchboard, but assigned a different set of evidence codes to reflect the lack of biochemical experiments. The FUS3/STE12/FUS1 triplet highlights the need to separate biochemical and genetic evidence to study PTM-dependent regulation because genetic evidence can often mask more complex mechanisms. The basic knowledge that a modifying enzyme activates or represses a TF cannot be assumed to involve a PTM in all cases.
DISCUSSION
Here, we report the development of a new kind of database—PTM-Switchboard—to catalog TF-PTM mediated regulation of gene transcription. As illustrated by the cases of CREB and the FUS3/STE12/FUS1 triplet, curating known MFG triplets requires an expert examination of literature. We have curated over 100 complete MFG triplets, thus establishing the groundwork for continued efforts. The database provides a focused and structured platform to leverage other regulatory network and signaling pathway resources for this multi-faceted task. In the future, we plan to extend this framework to other organisms and explore modeling and text-mining approaches to expand the collection. We have also made an effort to facilitate community involvement by allowing user submission of additional MFG triplets.
The first version of PTM-Switchboard is meant as the starting point towards a comprehensive database, but can also serve several more immediate purposes. For the molecular biologist studying a particular gene or pathway, PTM-Switchboard is available as a repository of structured MFG triplet data that is otherwise tedious to extract from the literature. Indeed, specific protein and transcription factor databases such as UniProt (34), TRANSFAC (35), and dbPTM (36) include information relevant to MFG-triplet. However, integration of these resources to extract consistent information needed for MFG-triplet ascertainment is not straightforward. For the computational biologist, the entirety of the database (or a subset filtered by an appropriate evidence code) can serve as an ideal gold set for training and testing models of three-way regulatory interactions (10,11). Moreover, the computational predictions can be conveniently compiled in PTM-Switchboard and made accessible to molecular biologists for experimental validation.
The collection is immediately useful to researchers interested in the approximately two-dozen modifying enzymes and transcription factors included in the first version, or as a resource for studying the regulation of over 100 target genes. PTM-Switchboard provides extensive connectivity to other data repositories, making it an ideal portal for researchers studying transcriptional regulation or cell signaling in Saccharomyces cerevisiae. Furthermore, transcriptional genomics studies typically reveal TFs potentially mediating the transcriptional control in a biological process of interest, for instance through motif enrichment analyses. The knowledge catalogued in PTM-Switchboard will be immediately helpful in designing further validation experiments and to place these experimental observations in a broader context of cell signaling.
Several approaches have been proposed to infer regulatory triplets based on expression data (10–12). The experimentally validated cases catalogued in PTM-Switchboard provide an ideal ‘gold set’ for benchmarking the performance of these methods on the more specific task of identifying MFG triplets. Furthermore, investigation of transcriptional networks based entirely on genomic and transcriptomic information, as is the current practice, is limited. This collection is designed to encourage more integrative computational approaches based on posttranscriptional data sources. By cataloging known instances of TF-PTM mediated regulation of gene transcription, PTM-Switchboard bridges the current resources in the fields of cell signaling and transcriptional regulation to facilitate a broader understanding of regulatory networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (R21GM078203, R01GM085226, T32HG000046). Funding for open access charge: R21GM078203.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
The authors would like to thank Greg Gonye for helpful feedback on the article.
REFERENCES
- 1.Creasy DM, Cottrell JS. Unimod: protein modifications for mass spectrometry. Proteomics. 2004;4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- 2.Garavelli JS. The RESID Database of protein modifications as a resource and annotation tool. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- 3.Khidekel N, Hsieh-Wilson LC. A ‘molecular switchboard’—covalent modifications to proteins and their impact on transcription. Org. Biomol. Chem. 2004;2:1–7. doi: 10.1039/b312466e. [DOI] [PubMed] [Google Scholar]
- 4.Brivanlou AH, Darnell JE. Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 5.Tootle TL, Rebay I. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays. 2005;27:285–298. doi: 10.1002/bies.20198. [DOI] [PubMed] [Google Scholar]
- 6.Freiman RN, Tjian R. Regulating the regulators: lysine modifications make their mark. Cell. 2003;112:11–17. doi: 10.1016/s0092-8674(02)01278-3. [DOI] [PubMed] [Google Scholar]
- 7.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 8.Holmberg CI, Tran SEF, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci. 2002;27:619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin WG. Proline hydroxylation and gene expression. Ann. Rev. Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Banerjee N, Margolin A, Nemenman I, Califano A. In Proceedings of the 10th Annual International Conference on Research in Computational Molecular Biology. Springer; 2005. Genome-wide discovery of modulators of transcriptional interactions in human B lymphocytes. [Google Scholar]
- 11.Zhang J, Ji Y, Zhang L. Extracting three-way gene interactions from microarray data. Bioinformatics. 2007;23:2903–2909. doi: 10.1093/bioinformatics/btm482. [DOI] [PubMed] [Google Scholar]
- 12.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gough NR. Science's signal transduction knowledge environment: the connections maps database. Ann. N. Y. Acad. Sci. 2002;971:585–587. doi: 10.1111/j.1749-6632.2002.tb04532.x. [DOI] [PubMed] [Google Scholar]
- 15.Choi C, Krull M, Kel A, Kel-Margoulis O, Pistor S, Potapov A, Voss N, Wingender E. TRANSPATH-a high quality database focused on signal transduction. Comp. Funct. Genomics. 2004;5:163–168. doi: 10.1002/cfg.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan X, Hu ZZ, Wu HT, Torii M, Narayanaswamy M, Ravikumar KE, Vijay-Shanker K, Wu CH. An online literature mining tool for protein phosphorylation. Bioinformatics. 2006;22:1668–1669. doi: 10.1093/bioinformatics/btl159. [DOI] [PubMed] [Google Scholar]
- 17.Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22:645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- 18.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 19.Linding R, Jensen LJ, Ostheimer GJ, van Vugt M.ATM, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rep M, Proft M, Remize F, Tamás M, Serrano R, Thevelein JM, Hohmann S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 2001;40:1067–1083. doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- 21.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 24.Pascual-Ahuir A, Struhl K, Proft M. Genome-wide location analysis of the stress-activated MAP kinase Hog1 in yeast. Methods. 2006;40:272–278. doi: 10.1016/j.ymeth.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Bardwell L, Cook JG, Voora D, Baggott DM, Martinez AR, Thorner J. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagen DC, McCaffrey G, Sprague GF. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:2952–2961. doi: 10.1128/mcb.11.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Ann. Rev. Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 30.Crosby JA, Konopka JB, Fields S. Constitutive activation of the Saccharomyces cerevisiae transcriptional regulator Ste12p by mutations at the amino-terminus. Yeast. 2000;16:1365–1375. doi: 10.1002/1097-0061(200011)16:15<1365::AID-YEA630>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 31.Elion EA, Satterberg B, Kranz JE. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung W, Olson KA, Breitkreutz A, Sadowski I. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur. J. Biochem. 1997;245:241–251. doi: 10.1111/j.1432-1033.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- 33.Song D, Dolan JW, Yuan YL, Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- 34.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wingender E, Dietze P, Karas H, Knüppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee T, Huang H, Hung J, Huang H, Yang Y, Wang T. dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]