Abstract
TarBase5.0 is a database which houses a manually curated collection of experimentally supported microRNA (miRNA) targets in several animal species of central scientific interest, plants and viruses. MiRNAs are small non-coding RNA molecules that exhibit an inhibitory effect on gene expression, interfering with the stability and translational efficiency of the targeted mature messenger RNAs. Even though several computational programs exist to predict miRNA targets, there is a need for a comprehensive collection and description of miRNA targets with experimental support. Here we introduce a substantially extended version of this resource. The current version includes more than 1300 experimentally supported targets. Each target site is described by the miRNA that binds it, the gene in which it occurs, the nature of the experiments that were conducted to test it, the sufficiency of the site to induce translational repression and/or cleavage, and the paper from which all these data were extracted. Additionally, the database is functionally linked to several other relevant and useful databases such as Ensembl, Hugo, UCSC and SwissProt. The TarBase5.0 database can be queried or downloaded from http://microrna.gr/tarbase.
INTRODUCTION
Mature microRNA (miRNA) molecules are approximately 22-nucleotide-long single-stranded RNAs that generally repress the expression of protein coding genes. Specifically, they preferentially bind to 3′ untranslated regions (UTRs) of messenger RNAs (mRNAs) and interfere with their stability and translational efficiency (1,2).
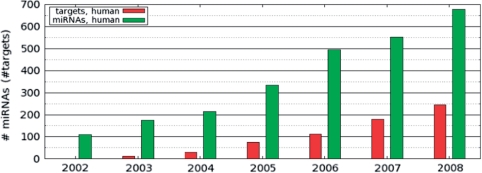
The first miRNAs and their target genes were identified via classical forward genetic techniques in 1993, but it was not until 2001 that many more miRNAs were discovered experimentally and found to be abundant and widespread (3–5). Since then there has been a dramatic growth in the number of annotated miRNAs (Figure 1).
Figure 1.
The growth of the human miRNA genes in mirBase database and the growth of the human experimentally determined miRNA target interactions in TarBase.
A crucial aspect of the functional analysis of miRNAs is the annotation of their protein-coding targets. A number of computational algorithms have been developed for the prediction of such targets (6). Although these programs are very important to guide wet lab experiments, they still lack in sensitivity and specificity (7,8).
In parallel, and as support for these programs, a number of experimental procedures have been developed to provide indirect or direct support for predicted miRNA–target interactions and results from a growing number of such experiments have been published (Figure 1).
The need for a systematic documentation of such experimentally supported targets was covered by the first version of TarBase (9). In 2006, the database recorded over 550 entries with miRNA–target interactions in human, mouse, fruit fly, worm and zebrafish. Here we present a substantially updated and extended version of this database, TarBase5.0, documenting over 1300 entries. The new database now contains all of the information included in the previous version, plus:
specific cells lines (if any) used in the experiments,
cell-type-specific expression of the gene product and its potential involvement in carcinogenesis,
differential expression of miRNAs in specific tissues,
developmental or pathological events that a specific miRNA is involved in and any annotated types of miRNA-related mis-regulation in those events,
HGNC Symbols as defined by HUGO (in order to provide consistent gene naming).
Additionally, both the underlying SQL database and the user interface have been extensively redesigned with several added external links such as a direct link to the paper's abstract on the PubMed site.
The TarBase5.0 database can be directly accessed from the http://microrna.gr/tarbase web page.
METHOD AND RESULTS
TarBase5.0 contains data extracted from a total of 203 scientific papers resulting in 1333 entries describing a regulatory interaction between a miRNA and a target 3′ UTR (summarized in Table 1).
Table 1.
A list of all TarBase5.0 entries
| Organism | Number of papers | Number of entries | Microarray data | pSILAC data |
|---|---|---|---|---|
| Homo sapiens | 110 | 285 | 328 | 474 |
| Mus musculus | 28 | 105 | 13 | – |
| D. melanogaster | 23 | 77 | – | – |
| C. elegans | 18 | 14 | – | – |
| Plants | 21 | 30 | – | – |
| Danio rerio | 1 | 1 | – | – |
| Rat | 2 | 2 | – | – |
| Total | 203 | 514 | 341 | 474 |
The TarBase5.0 data set contains miRNA targets that tested either positive (induces target gene repression) or negative (no influence on target gene expression). For each experiment with a positive outcome the target site is described by the miRNA that binds it, the gene in which it occurs, the nature of the experiments that were conducted to test it, the sufficiency of the site to induce translational repression and/or cleavage, and the paper from which all these data is extracted. Additionally, for each miRNA and protein-coding gene, the database contains links to several other relevant and useful databases such as Ensembl (10), Hugo (11), UCSC genome browser (12) and SwissProt (13).
There are a number of direct and indirect experimental procedures that have been developed to test a possible miRNA–mRNA interaction. The entries in Tarbase5.0 are classified into four categories: TRUE or FALSE in the cases where an assay provides direct experimental evidence, or MICROARRAY and/or pSILAC in the cases that present only indirect evidence from high-throughput techniques to measure miRNA-mediated global transcriptomic or proteomic changes. All of these approaches make use of technology for miRNA knock down or overexpression. To overexpress a miRNA, expression constructs can be engineered using the mature miRNA, the precursor (hairpin) miRNA, or the pri-miRNA sequence for transfection into in vitro or in vivo transformed cells. Also, silencing of a specific miRNA can be accomplished by introducing chemically modified oligonucleotides that are perfectly complimentary to the mature miRNA (antagomirs) (14). These methods for modifying miRNA expression allow for several types of follow-up techniques to quantify and interpret differences in target gene expression. Below we provide a more detailed description of each of the four categories:
TRUE or FALSE: The most commonly used method for providing direct experimental evidence is the reporter gene assay. In its simplest form, an expression vector containing a reporter gene [i.e. Luciferase or Green Fluorescent Protein (GFP)] is first modified by cloning the predicted target 3′UTR downstream of the reporter gene, and then transfected into a cell line of interest in the absence and presence of the cognate miRNA. Despite the general utility of this approach to assay for 3′UTR-mediated effects on reporter protein expression, it is not informative for the precise location of the miRNA response element (MRE) or number of miRNA target sites in the 3′UTR. Integration of the reporter gene assay with site directed mutagenesis of the predicted MRE (and, further, restoring the complementarity of the miRNA–MRE interaction by mutating the mature miRNA sequence) yields a much more specific and direct result. To measure effects on reporter mRNA levels, the most commonly applied technique is quantitative RT-PCR (qRT-PCR). Measuring effects on both protein and mRNA levels can help provide information about the mode of miRNA-mediated silencing: mRNA translational repression or immediate RISC-mediated mRNA cleavage and degradation. A miRNA–MRE interaction is reported as TRUE or FALSE based on the results of the reporter gene assay.
MICROARRAY and/or pSILAC: These high-throughput approaches measure global changes in the transcriptome (15) or proteome (8,16) given the presence or absence of a miRNA. Despite their power for large-scale analysis, these techniques only provide indirect evidence about a miRNA's targets since it is not possible to distinguish between primary direct targets and secondary indirect targets. Other high-throughput methods like degradome sequencing (17,18) are also immensely useful but only in the scenarios where a miRNA induces RISC-mediated mRNA cleavage.
In order to facilitate user interaction, the query function is divided into several functionally related subgroups. The initial screen of the TarBase5.0 user interface allows users to query based on miRNA, gene and organism. For more advanced queries, the user can utilize the extended query options. In this case, the search menus are arranged into four functionally related groups.
The first group contains the fields with information about the miRNA–target interaction: the validity of the interaction (field ‘Support Type’, either true or false), the function of the interaction which can be either translational repression or mRNA cleavage (field ‘DataType’), the sufficiency of a single target site to exert the specific function (field ‘S_S_S’) and the number of miRNA response elements present in the specific UTR (field ‘MRE’).
The second group contains the fields that refer to the experimental methods that led to the reported result. The field ‘Direct Support’ refers to experimental procedures that provide direct evidence regarding the miRNA–target interaction (i.e. reporter gene assays) while ‘Indirect Support’ refers to experimental procedures that provide more global, system-wide miRNA-mediated effects (i.e. microarrays).
The third group corresponds to biological properties of the miRNA or target gene: biological functions (field ‘Protein Type’), specific expression profiles (field ‘miRNA Expression’) or the physiological processes in which this interaction is involved (field ‘Event or Pathology’). The fourth and final group contains some general query features such as the scientific paper (searchable by Author or PMID).
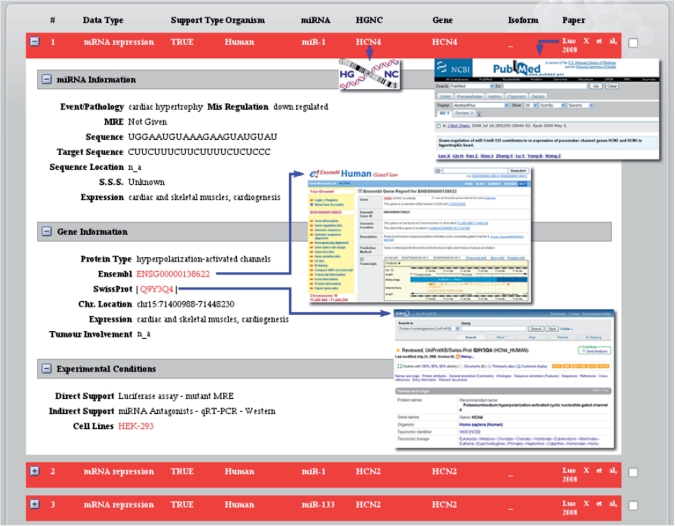
The results are presented in a similar format as the query fields. By default, the results screen (Figure 2) shows only the repression type, the miRNA identifier, the target gene identified by the HGNC symbol (if it is a human gene), the common gene name, the Refseq isoform id (particularly relevant in cases of gene variants or SNP haplotypes), the affected biological processes and the paper containing the information presented. Users can opt to view more detailed information by clicking on the ‘+’ box so that the expanded results view is opened (Figure 2). The additional information is divided into three categories: miRNA information, gene information and experimental conditions.
Figure 2.
Example of a result screen for a TarBase query. The context-specific links to other resources are indicated by the blue arrows.
The ‘miRNA information’ category contains the properties of the specific miRNA such as the miRNA's sequence [extracted from miRBase (19)], the number and sequences of the MREs, their locations within the gene's 3′UTR, and the affected tissues (extracted from the paper). The ‘Gene information’ category gathers mostly biological properties of the target gene like the protein type, Ensembl and SwissProt IDs and chromosome location, providing direct links to Ensembl, SwissProt and the UCSC browser respectively. Moreover, expression profiles and tumor involvement are also provided for human genes (information extracted from the Ensembl eGenetics database). Finally, the ‘Experimental conditions’ category provides the nature of the direct or indirect evidence for the miRNA–target gene interaction. The cell lines used to carry out the specific experiment are also presented in order to render the experimental conditions more complete and reproducible.
CONCLUSION
Even though several computational programs exist to predict miRNA targets, the necessity for a systematic collection and description of miRNA targets with experimental support led to the development of TarBase. The continuously expanding number of known and newly identified miRNAs and their targets, combined with their central role in biological systems, renders this field particularly dependent on centralized information that is accurate, up-to-date, comprehensive and easy to browse or download. In order to satisfy these requirements, we have made extensive updates and modifications and present the new version of the database, TarBase5.0.
AVAILABILITY
TarBase is freely available at http://microrna.gr/tarbase. The TarBase data files can be freely downloaded and used according to the GNU Public License. The relevant literature is reviewed for new entries and the database is updated quarterly.
FUNDING
Funding for open access charge: Aristeia Award from General Secretary Research and Technology, Greece.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 2.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 7.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 8.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 9.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangan ME, Williams JM, Lathe SM, Karolchik D, Lathe W.C., 3rd. UCSC Genome Browser: Deep support for molecular biomedical research. Biotechnol. Annu. Rev. 2008;14:63–108. doi: 10.1016/S1387-2656(08)00003-3. [DOI] [PubMed] [Google Scholar]
- 13.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–6. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]