Abstract
MachiBase (http://machibase.gi.k.u-tokyo.ac.jp/) provides a comprehensive and freely accessible resource regarding Drosophila melanogaster 5′-end mRNA transcription at different developmental states, supporting studies on the variabilities of promoter transcriptional activities and gene-expression profiles in the fruitfly. The data were generated in conjunction with the recently developed high-throughput genome sequencer Illumina/Solexa using a newly developed 5′-end mRNA collection method.
INTRODUCTION
Characterization of the complete repertoire of expressed messenger RNA (mRNA) is central to the functional analysis of a genome. To date, several studies have been undertaken to achieve a better understanding of the Drosophila melanogaster genome (1–4). The technical approaches used in these studies included in-depth, full-length cDNA cloning and tiling microarrays. However, despite the absence of prior knowledge of the locations of previously identified genes, the 5′-end SAGE (5) method has demonstrated efficacy in cataloging high numbers of expressed genes. Following the simple modification of adopting the recently developed high-throughput genome sequencer Illumina/Solexa, 5′-end SAGE has become a potent tool for elucidating transcriptional mechanisms. To achieve a deeper insight into transcriptional activity, we collected approximately 25 million 25–27 nt 5′-end mRNA tags from the embryos, larvae, young males, young females, old males, old females and S2 (culture cell line) of D. melanogaster with high mechanical reproducibility. After aligning these tags to unique positions in the fly genome while allowing three mismatches, 2.87–4.05 million uniquely mapped tags were amassed for each of the seven samples. These data constitute the most substantial transcriptional start site (TSS) and gene-expression database for D. melanogaster currently available.
MachiBase is designed to assist fly biologists in their analyses of gene expression and in placing expression data in the context of functional genomics through genomic orientation. Thus, information on differentially expressed genes can be accessed by either inputting the gene name as a keyword or selecting a chromosomal location. Aside from providing information on gene expression, these data constitute a potent resource for analyses of transcriptional regulation. The core promoter, which is the region surrounding the TSS of a gene required for recruitment of the transcription apparatus, warrants analysis. However, TSSs and core promoters have previously been identified on a gene-by-gene basis. With the help of this database, biologists can explain transcriptional initiation mechanisms by combining additional information on chromatin structure and DNA methylation. In addition, these data allow accurate predictions of gene structures, particularly of the 5′-untranslated region (5′-UTR).
METHODS
The newly developed 5′-end mRNA collection method extends the range of the original 5′-end SAGE technique developed by Hashimoto et al. (5). This method initially profiles 25–27 nt tags using a novel strategy that incorporates the oligo-capping method (6). The 5′-end tags are then ligated directly to the Illumina/Solexa linker, to prepare for sequencing with the Illumina/Solexa system. Prior to construction of the Illumina/Solexa libraries, we confirmed the integrity of the cDNA using the Agilent 2100 Bioanalyser.
Collection of numerous 5′-end tags from seven libraries and testing the reproducibility of the method used
To characterize the transcriptional activity patterns of the D. melanogaster genome, we collected 25–27 nt 5′-end mRNA tags from embryos, larvae, young males, young females, old males, old females and the S2 cell line. Table 1 presents the results of this process. The second column shows more than five million raw tags collected from each of the seven libraries. As most of these tags were redundant, they were grouped into non-redundant representative tags, the statistics for which are shown in the third column. Each non-redundant tag represents a duplicated occurrence and is therefore associated with its frequency, i.e. the number of times that it occurs.
Table1.
Statistical analysis of collected tags and identified TSSs from the seven libraries
| Library | Number of raw, redundant tags (A) | Number of raw, non-redundant tags | Number of uniquely aligned, redundant tags (B) | B/A (%) |
|---|---|---|---|---|
| Embryo | 5 620 821 | 2 123 688 | 3 321 095 | 59.1 |
| Larvae | 6 711 841 | 2 231 078 | 3 556 021 | 53.0 |
| Young male | 11 349 959 | 7 859 645 | 3 721 539 | 32.8 |
| Young female | 6 882 149 | 3 398 754 | 2 875 247 | 41.8 |
| Old male | 7 198 682 | 3 442 886 | 3 873 078 | 53.8 |
| Old female | 6 787 420 | 3 258 523 | 3 683 536 | 54.3 |
| S2 | 7 214 104 | 2 803 878 | 4 052 965 | 56.2 |
| Total | 51 764 976 | 25 083 481 | 48.5 |
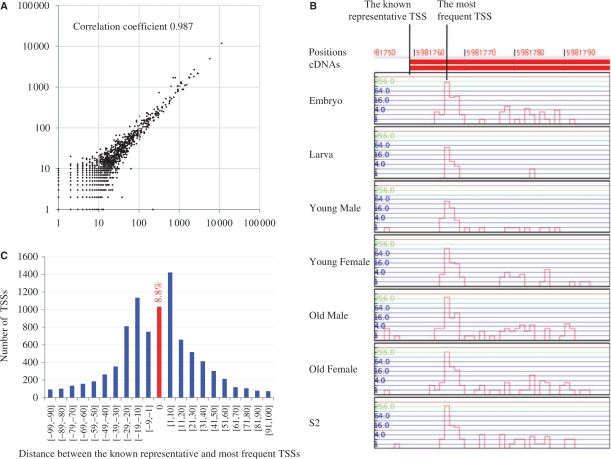
The frequency is expected to be reproducible, in that the frequency of each non-redundant tag is proportional to the total number of tag occurrences in independent experiments. To test for reproducibility, we performed an additional collection of 5′-end tags from the same young female Drosophila library. Figure 1A reveals a strong correlation between the two independent experiments. Furthermore, in a comparison with a quantitative PCR analysis, the employed method has been validated as a means to quantify the expression level of a transcript as the number of 5′-end tags (7).
Figure 1.
Statistical analysis of the TSS information. (A) A dot represents one non-redundant 5′-end tag, such that the values on the x-axis and y-axis indicate the frequencies of the focal tags in the respective experiments. (B) A case in which the known representative TSS is not consistent with the most frequent TSS. Note that the most abundant TSSs coincide across the seven different libraries. (C) The distribution of distances between the known representative TSSs and most frequent TSSs. Overall, 1033 (8.8%) of the 11 725 known representative TSSs coincide with the most frequent TSSs.
Identification of transcription start sites by millions of 5′-end tags
For the identification of TSSs, non-redundant tags were aligned to the genome of D. melanogaster (R5.3) in FlyBase (8). We observed that 5′-end tags tended to contain read errors, especially towards their termini. To correct these read errors, the tags were aligned to the genome while allowing, at most, three mismatches. The efficient mapping of millions of tags was an issue that needed to be resolved. We developed and used a parallel version of BLAT (9), which operates on massive parallel clusters. Another major technical issue involved the fact that a single 5′-end tag could be mapped to multiple locations, making it difficult to determine the original location of the tag. To eliminate false-positive positional data, these ambiguous tags were simply excluded from our analysis, so that only uniquely aligned tags were considered. A tag was considered to be uniquely aligned if, for a non-negative number k (⩽3), the tag was mapped to a unique location with ⩽k mismatches, although it could be mapped to multiple positions with more than k mismatches. The number of uniquely aligned and redundant tags in each library, and their ratios to the number of raw redundant tags, are shown in the fourth and fifth columns of Table 1, respectively. A uniquely aligned 5′-end tag identified a TSS in the genome. Distinct tags could be mapped to the same TSS, since the alignment step tolerated mismatches and replaced erroneous nucleotides with the correct nucleotides in the genome. From all seven libraries, a total of 25 083 481 tags were mapped to unique locations, thereby identifying 1 773 851 TSSs; the data breakdown in terms of chromosomes is presented in Table 2.
Table 2.
Breakdown of the uniquely aligned, redundant tags in terms of chromosomes
| Chromosome | Number of TSSs |
|---|---|
| 2L | 323 720 |
| 2L heterochromatin | 406 |
| 2R | 369 405 |
| 2R heterochromatin | 1801 |
| 3L | 320 952 |
| 3L heterochromatin | 1930 |
| 3R | 442 109 |
| 3R heterochromatin | 1691 |
| 4 | 10 975 |
| U | 3506 |
| U extra | 9398 |
| X | 286 427 |
| X heterochromatin | 1305 |
| Y heterochromatin | 93 |
| Mitochondria | 133 |
| Total | 1 773 851 |
Discrepancy between the known representative TSS and the most frequent TSS
In attempting genome annotation, it is usual to choose the longest cDNA sequence in a specific locus to define the representative cDNA. To examine the level of agreement between the newly collected 5′-end tags and the known representative cDNA sequences, we calculated how many of the uniquely aligned, redundant tags were located in the promoters and 5′-UTRs of the representative sequences, and found 96.2% of the 5′-end tags in the UTR regions (Table 3). Figure 1B illustrates the 5′-end tag expression patterns surrounding a representative TSS in the seven libraries. It was intriguing to observe that the representative TSS was not necessarily the most frequent TSS, but that another TSS slightly downstream of the representative was the most abundant, which motivated us to examine this discrepancy. We calculated the distances between the representative TSSs and the most frequent TSSs in the promoters and 5′-UTRs of the 11 725 longest cDNA sequences in FlyBase. Figure 1C shows the numbers of representative TSSs in terms of distances, highlighting that only 1033 (8.8%) of the 11 725 known representative TSSs were the most frequent TSSs. Our analysis indicates that the common practice of selecting the longest cDNA sequence as the representative one needs to be revised, and demonstrates the efficacy of 5′-end tag collection for detecting the most abundant TSS as an alternative to the representative TSS.
Table 3.
Ratios of uniquely aligned, redundant tags located in the promoters and 5′-UTR of the representative sequences
| 5′UTR + promoters (500 bp upstream) | Uniquely aligned, redundant tags | ||
|---|---|---|---|
| Embryo | 3 212 489 | (96.7%) | 3 321 095 |
| Larvae | 3 350 688 | (94.2%) | 3 556 021 |
| Young male | 3 528 146 | (94.8%) | 3 721 539 |
| Young female | 2 757 521 | (95.9%) | 2 875 247 |
| Old male | 3 750 286 | (96.8%) | 3 873 078 |
| Old female | 3 583 360 | (97.3%) | 3 683 536 |
| S2 | 3 958 105 | (97.7%) | 4 052 965 |
| Total | 24 140 595 | (96.2%) | 25 083 481 |
Database features and applications
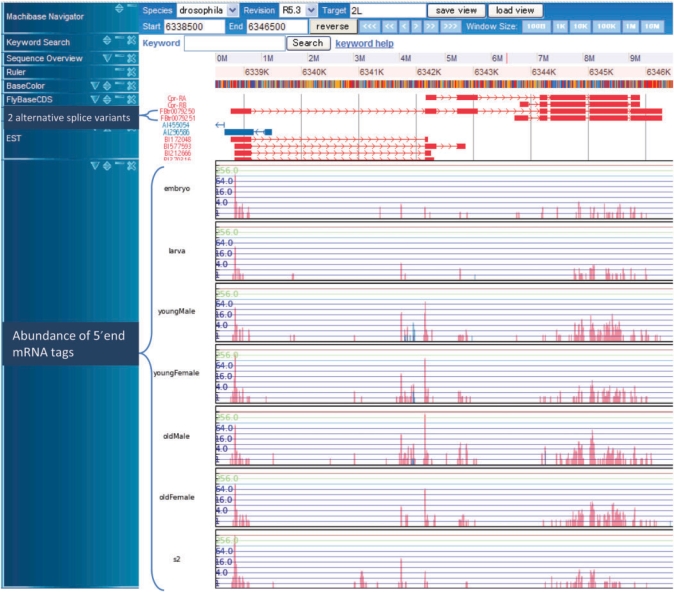
We visualized the numbers of 5′-end tags for each position in a vertical bar (Figure 2). This arrangement of 5′-end data provides an insight into fly transcription, in combination with other annotated genomic information. In the MachiBase database server, users can browse the TSSs and frequencies of individual genes by querying the FlyBase gene ID, FlyBase transcription ID, etc. In addition to the gene-specific view, it is also possible to generate an overview of all the expressed transcripts for an assigned position on a chromosome. Furthermore, all these genes are linked with the FlyBase annotation server, which contains Gene Ontology (GO), orthologue information, etc. In addition to revealing differentially expressed genes, genome-wide TSS discovery is a valuable resource for biologists studying flies. This high-throughput study has revealed a surprisingly large number of novel genic (intron–exon regions) and intergenic TSSs, which has prompted a rethink of the relationships between gene transcription and promoter architecture. For example, if we display the location (2L: 2 391 450–2 391 850) by inputting 2L into the ‘Target’ box, 2 391 450 into the ‘Start’ box, 2 391 850 into the ‘End’ box, we can see the existence of an a new transcript supported by a significant number of 5′-end tags in the un-annotated intergenic region. Thus, the precise locations of the TSSs enable an in-depth analysis of cis-acting elements that are bound by transcription factors. This data resource provides a starting point for elucidating novel molecular details of transcription by reliably integrating TSS location data with related functional data, such as histone methylation and acetylation states (10,11), the positions of nucleosomes (12–14) and the occupancy of transcription factor binding sites (15), each of which, as features, can now be examined on a genome-wide basis.
Figure 2.
Snapshot of the MachiBase genome browser. The frequencies of the 5′-end mRNA tags mapped to individual positions on the fruitfly genome in the seven libraries are displayed as histograms in the bottom seven tracks. In the histograms, the vertical bars in log scale indicate the numbers of 5′-end mRNA tags aligned to each position on the x-axis. The upper track shows the exon–intron structures of two alternative splice variants. Observe that the peaks for the 5′-end tags are around the 5′-end of the longer splice variant in all of the seven libraries. In addition, note that many 5′-end tags are expressed from the second, second-to-last, and last exons in four adult samples (young/old and male/female).
DISCUSSION
The vast transcriptional datasets have been used to characterize differentially expressed genes, especially in relation to age and sexual development. Using these datasets, we have confirmed that the representative TSSs, the abundantly expressed TSSa flanking FlyBase-annotated TSSs, differ from many of the known FlyBase-annotated TSSs. It has become evident that the rules for start site selection are fundamentally different for different promoters, and large-scale studies have given us the tools to partition promoters into functional classes with respect to TSS information in future studies. As a novel and high-quality data resource, MachiBase is a valuable tool for experimental biologists who are working on D. melanogaster. In future, we will empower this database with various annotated data on the fly genome.
FUNDING
Scientific Research on Priority Areas (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan partially; Bioinformatics Research and Development (BIRD); the Japan Science and Technology Agency (JST). Funding for open access charge: JST.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Computational time was provided by the Information Technology Center and the Human Genome Center, at the University of Tokyo. All 5′-end sequence data are deposited at NCBI Short Read Archive under the accession number SRA002200.
REFERENCES
- 1.Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- 2.Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, et al. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 4.Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto S, Suzuki Y, Kasai Y, Morohoshi K, Yamada T, Sese J, Morishita S, Sugano S, Matsushima K. 5′-end SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- 6.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto S, Qu W, Budrul A, Ogoshi K, Nakatani Y, Lee Y, Ogawa M, Ametani A, Suzuki Y, Sugano S, et al. High-resolution analysis of the 5′-end transcriptome using a next generation DNA sequencer. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drysdale RA, Crosby MA. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol. Cell Biol. 2006;26:6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 2005;24:2906–2918. doi: 10.1038/sj.emboj.7600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishida H, Suzuki T, Kondo S, Miura H, Fujimura YI, Hayashizaki Y. Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome Res. 2006;14:203–211. doi: 10.1007/s10577-006-1036-7. [DOI] [PubMed] [Google Scholar]
- 14.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu YT, Weng ZP, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]