Abstract
The EST division of GenBank, dbEST, is widely used in many applications such as gene discovery and verification of exon–intron structure. However, the use of EST sequences in the dbEST libraries is often hampered by inconsistent terminology used to describe the library sources and by the presence of contaminated sequences. Here, we describe CleanEST, a novel database server that classified dbEST libraries and removes contaminants. We classified all dbEST libraries according to species and sequencing center. In addition, we further classified human EST libraries by anatomical and pathological systems according to eVOC ontologies. For each dbEST library, we provide two different cleansed sequences: ‘pre-cleansed’ and ‘user-cleansed’. To generate pre-cleansed sequences, we cleansed sequences in dbEST by alignment of EST sequences against well-known contamination sources: UniVec, Escherichia coli, mitochondria and chloroplast (for plant). To provide user-cleansed sequences, we built an automatic user-cleansing pipeline, in which sequences of a user-selected library are cleansed on-the-fly according to user-selected options. The server is available at http://cleanest.kobic.re.kr/ and the database is updated monthly.
INTRODUCTION
Expressed sequence tag (EST) sequences are generated by single-pass DNA sequencing of clones randomly selected from cDNA libraries and represent partial descriptions of the transcribed portions of genomes (1). EST sequences are widely used for the rapid and cost-effective discovery of new genes, verification of the exon-intron structure of predicted genes, and as resources for gene mapping and cDNA array construction (2). The most extensive resource of EST data is the EST division of GenBank, dbEST, which is hosted by The National Center for Biotechnology Information (NCBI) (3). Sequences in dbEST have been submitted by various EST sequencing projects and are freely available. Since 1992, when the first EST data appeared in GenBank, the number of EST sequences has dramatically increased. As of July 2008, dbEST contained more than 54 million sequences (from about 22 000 libraries) and accounts for nearly 62% of all GenBank entries (4).
The sequences submitted to dbEST were created from thousands of different cDNA libraries and may have originated from whole organs (for example, human brain and liver), specialized tissues, or individual cells (5). Some libraries were developed to compare transcripts from different developmental stages; others were developed to highlight differences in gene expression between normal and transformed cells. However, the researchers who submit EST sequences can use highly inconsistent terminology to describe library sources and this can impair effective searching by computers and individuals. For example, if a user is searching for all cDNA libraries generated from lymphoid tissue, he might use several equivalent terms (for example, ‘lymph’ and ‘tonsil’) for the searching. Searching with only one of these equivalent terms might produce an incomplete result. Therefore, to obtain all the cDNA libraries created from a tissue or cell line of interest, a controlled vocabulary is necessary.
Another obstacle in the identification of encoded genes from dbEST is the existence of contamination that was introduced during construction of cDNA libraries and sequencing of inserted cDNA (6). EST data are produced from various libraries by different sequencing technologies. The most common contaminants are vector/linker sequences, library host genomes (usually Escherichia coli), and sequences from cytoplasmic organelles such as mitochondria (7). These types of artifacts can cause erroneous clustering and assembly during reconstruction of putative transcripts and may ultimately lead to inaccurate gene annotation. Such contaminated sequences must be thoroughly cleansed before analysis (8). ‘Cleansing before using’ is essential for all EST analyses with dbEST or user-specific EST datasets (9). Although some libraries in dbEST are reported to be highly contaminated (10), there are few studies of the artifacts in dbEST libraries (11). We know of no database server that provides cleansed dbEST libraries.
Here, we present a web-based database server, CleanEST, to provide cleansed EST sequences of classified dbEST libraries. To illustrate its function, we classified EST sequences that were downloaded from dbEST according to organism, sequencing center and eVOC ontology (12) for human libraries. We compared the EST sequences of the libraries against major contamination databases and trimmed or completely discarded contaminated sequences. Finally, we integrated them into a relational database that is accessed via web-based user interfaces.
DATABASE CONTENTS AND METHODS
Dataset
EST sequences were downloaded from dbEST at the NCBI GenBank FTP site (ftp://ftp.ncbi.nih.gov/genbank/), converted into FASTA-formatted sequences, and divided according to library names. As of July 2008, there were 22 457 libraries and 54 447 050 sequences. The number of sequences in each library varied from 1 to 541 852. The average length of sequences was 549 bases with a SD of 250.
Classification of dbEST libraries
All libraries were classified by organism and sequencing center. There were 1686 species and 2551 sequencing centers in dbEST. There are 8668 human libraries in dbEST, more than any other species. However, these human libraries were generated from various sources and there are significant differences in the terminology used to describe the library sources. Thus, we further categorized the human libraries by a set of structured and controlled terms from eVOC, ontologies with strict hierarchical structures that describe human anatomy, histology, development and pathology. We automatically assigned the human libraries to the Anatomy ontology and the Pathology ontology of eVOC. The assignments were obtained by a substring matching method, in which ‘organ’ or ‘tissue’ names in dbEST libraries must match anatomical and pathological terms of eVOC. In this procedure, 5608 (65%) of 8668 total human libraries were assigned to the Anatomy ontology and the remaining 3060 libraries were considered ‘unclassifiable’. In the Pathology ontology, we assigned 3075 (35%) human libraries to the ontology and the remaining 5593 libraries to ‘unclassifiable’. The use of the eVOC facilitates uniform queries across dbEST libraries and allows users to query at different levels.
Cleansing contaminated entries in EST sequences
For each dbEST library, CleanEST provides two different cleansed sequences: ‘pre-cleansed’ and ‘user-cleansed’. To provide pre-cleansed sequences, first, we obtained sequences of major contamination databases from the NCBI FTP site. The databases used were the UniVec database (for vector/linker), the Escherichia coli full genome sequence (for cloning host) and the RefSeq (13) mitochondrial genome sequences (for organelle) and chloroplast (14) genome sequences (for plant organelle). Second, EST sequences in each library were compared against these three database sequences and contaminated regions were masked. This was performed using cross_match (15) program, an implementation of the Smith–Waterman (16) algorithm. The minmatch and minscore parameters of the cross_match were set at 20 and 20 and all other parameters were kept at default values. Finally, masked EST sequences were either trimmed or discarded using our Perl script trimming tool. If masked regions commenced within 100 bases of the 5′- or 3′-ends, they were trimmed. EST sequences with internally located masked regions were discarded because we could not determine which part was from a cDNA transcript or because such sequences might be chimeras. After pre-cleansing, EST sequences shorter than 50 bases were discarded (17).
To provide user-cleansed sequences, CleanEST uses an automatic user-cleansing pipeline, in which sequences in a user-selected library are cleansed on-the-fly according to user-input options. This pipeline consists of highly reliable open-source tools and public databases. In the interface of the pipeline, user can select parameters of the cross_match program and contamination sources. In addition, the user can upload own contamination sources. After user-cleansing, users can download the cleansed sequences.
Contamination statistics
Our analysis of the pre-cleansed EST sequences indicates that 2 401 140 (4.8%) of the sequences in dbEST contained significant hits to at least one of the three contamination sources. Among these, 946 929 (1.9%), 379 293 (0.8%) and 1 438 645 (2.9%) partially or fully matched sequences in UniVec, E. coli and mitochondria, respectively. Of all contaminated sequences, 1 341 307 were trimmed and 1 059 833 were discarded. In this analysis, the three contamination sources may partly overlap because some vector sequences are from E. coli sequences, and some EST sequences could be matched with two or more contamination sources. The average length of a contaminated region was 147 bases. Even though most of the libraries had very low contamination rates (<10%), some were heavily contaminated (Table 1). Detailed contamination information is available on the CleanEST website.
Table 1.
Distribution of contaminated EST sequences in dbEST libraries
| Frequency of contaminated ESTs (%) | Nunber of libraries |
|---|---|
| >90 | 171 |
| 90–80 | 80 |
| 80–70 | 91 |
| 70–60 | 122 |
| 60–50 | 214 |
| 50–40 | 218 |
| 40–30 | 381 |
| 30–20 | 835 |
| 20–10 | 1963 |
| <10 | 18 382 |
Our analysis showed that the average matching lengths of contamination sequences from vector/linker, E. coli and mitochondria were 114 (19%), 116 (20%) and 382 (61%) bases, respectively. The numbers in parentheses indicate the average percentages of contaminated regions in contaminated sequences. For contamination by UniVec and E. coli, the contaminated regions accounted for <20% of the sequence length. For contamination by mitochondrial sequences, the matching average length was >60% of the corresponding sequences, about 3-fold larger than that of other contamination regions. This means that most mitochondrial transcripts were inserted into cloning vectors during cDNA library construction and were then sequenced as if they were genomic transcripts. In other words, mitochondrial transcripts replaced genomic transcripts. In contrast, the other contaminants were mostly appended into genomic transcripts and read as parts of genomic transcripts.
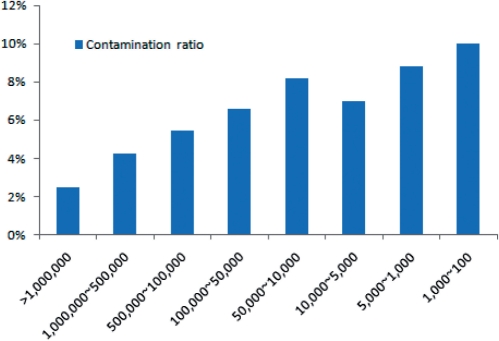
The number of sequences submitted by each sequencing center varied from 1 to 2 322 086. For this reason, we analyzed the contamination ratio of sequencing centers as a function of the number of submitted sequences. Thus, we divided sequencing centers into several groups according to the number of sequences deposited in dbEST and then calculated an average contamination ratio of each group. Interestingly, our analysis shows that the average contamination ratio is lower for centers that have submitted a larger number of sequences (Figure 1). Small sequencing centers (<10 000 ESTs) have more than double the contamination of large sequencing centers (>1 000 000 ESTs). This may be because large sequencing centers have more reliable sequence-cleansing facilities or because researchers in small sequencing centers are unaware of the problem of EST contamination when submitting an EST dataset to the EST repository.
Figure 1.
Distribution of average contamination ratio of sequencing centers. Sequencing centers were classified according to the number of their total sequences in dbEST and were calculated an average contamination ratio of each class. The x-axis represents the classes of sequencing centers and y-axis represents their contamination ratio. The average contamination ratio is lower for centers that have submitted a larger number of sequences. Small sequencing centers (<10 000 ESTs) have more than double the contamination of large sequencing centers (>1 000 000 ESTs).
Construction of web-based database server
The CleanEST database server is composed of a web interface and a MySQL database management system. The web interface is implemented in static HTML pages and Java Server Pages to allow database searching. We used MySQL to store the cleansed sequences and associated library classification information. The main web page has four types of search menus: organism, sequencing center, eVOC ontologies (for human libraries) and user sequences. In the organism and sequencing center menu, the user can select a species and sequencing center and then download all the EST sequences or list entire libraries that are associated with the selected item. In the query input fields of the two menus, we provide an ‘autocomplete’ function that shows the five best candidates related to the input words. Thus, the user can enter a query without knowing the exact spelling of the word.
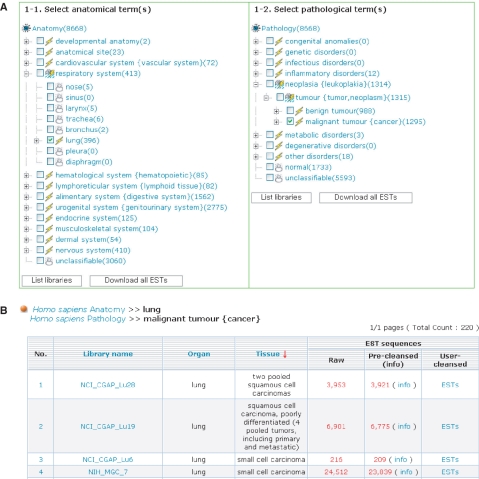
In the human library menu, the user can select anatomical or pathological terms from tree-shaped eVOC ontologies and then download relevant EST sequences as compressed files. Selecting anatomical and pathological terms allows the user to perform combination queries (Figure 2). For instance, a query of ‘lung’ in the Anatomy ontology returns all libraries related to liver and a query on ‘cancer’ in the Pathology ontology returns all libraries associated with tumor. The combination query returns the intersection (lung ∩ cancer) of these two libraries. In the result page, the user can sort the library list by clicking on organism, library name, organ or tissue. In the user sequence menu, the server provides a web interface to cleanse user-uploaded EST sequences.
Figure 2.
Screenshots of CleanEST showing the query, ‘lung AND cancer’. (A) Query of ‘lung’ in the Anatomy ontology and ‘cancer’ in the Pathology ontology of the eVOC search menu. (B) Library list showing the results of the query.
CONCLUDING REMARKS
We developed CleanEST, a database that contains raw and cleansed EST sequences of classified dbEST libraries. Our cleansing analysis showed that ∼4.8% of sequences in dbEST are contaminated by vector/linker, E. coli or mitochondrial sequences. We also found some libraries are heavily contaminated. Biological researchers who plan to use dbEST libraries in their research would benefit greatly from the use of CleanEST.
The cleansing procedure that we used is based on EST sequence alignments against commonly known contamination sequences and fully sequenced genomes. Thus, our cleansing procedure does not consider all types of contaminations, including intron sequences, other sequences present in immature eukaryotic mRNA and sequences from cloning vectors that are not in the UniVec database. Other contaminations are possible. Accordingly, some libraries might require further cleansing depending on the DNA sources and sequencing history of such libraries.
FUNDING
This work was supported by the Ministry of Education, Science and Technology (MEST) under grant number M10869030002-08N6903-00210 and the KRIBB Research Initiative Program. Funding for open access charge: MEST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nagaraj SH, Gasser RB, Ranganathan S. A hitchhiker's guide to expressed sequence tag (EST) analysis. Brief. Bioinform. 2007;8:6–21. doi: 10.1093/bib/bbl015. [DOI] [PubMed] [Google Scholar]
- 2.Lee B, Hong T, Byun SJ, Woo T, Choi YJ. ESTpass: a web-based server for processing and annotating expressed sequence tag (EST) sequences. Nucleic Acids Res. 2007;35:W159–W162. doi: 10.1093/nar/gkm369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for “expressed sequence tags”. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 4.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkinson J, Anthony A, Wasmuth J, Schmid R, Hedley A, Blaxter M. PartiGene—constructing partial genomes. Bioinformatics. 2004;20:1398–1404. doi: 10.1093/bioinformatics/bth101. [DOI] [PubMed] [Google Scholar]
- 6.Mao C, Cushman JC, May GD, Weller JW. ESTAP—an automated system for the analysis of EST data. Bioinformatics. 2003;19:1720–1722. doi: 10.1093/bioinformatics/btg205. [DOI] [PubMed] [Google Scholar]
- 7.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 8.D’Agostino N, Aversano M, Chiusano ML. ParPEST: a pipeline for EST data analysis based on parallel computing. BMC Bioinformatics. 2005;6(Suppl. 4):S9. doi: 10.1186/1471-2105-6-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorek R, Basechess O, Safer HM. Expressed sequence tags: clean before using. Correspondence re: Z. Wang et al., computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. 2003. Cancer Res., 63, 6996; author reply 6996–6997. [PubMed] [Google Scholar]
- 10.Sorek R, Safer HM. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 2003;31:1067–1074. doi: 10.1093/nar/gkg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seluja GA, Farmer A, McLeod M, Harger C, Schad PA. Establishing a method of vector contamination identification in database sequences. Bioinformatics. 1999;15:106–110. doi: 10.1093/bioinformatics/15.2.106. [DOI] [PubMed] [Google Scholar]
- 12.Kelso J, Visagie J, Theiler G, Christoffels A, Bardien S, Smedley D, Otgaar D, Greyling G, Jongeneel CV, McCarthy MI, et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui L, Veeraraghavan N, Richter A, Wall K, Jansen RK, Leebens-Mack J, Makalowska I, dePamphilis CW. ChloroplastDB: the Chloroplast Genome Database. Nucleic Acids Res. 2006;34:D692–D696. doi: 10.1093/nar/gkj055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 16.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 17.Negre V, Hotelier T, Volkoff AN, Gimenez S, Cousserans F, Mita K, Sabau X, Rocher J, Lopez-Ferber M, d’Alencon E, et al. SPODOBASE: an EST database for the lepidopteran crop pest Spodoptera. BMC Bioinformatics. 2006;7:322. doi: 10.1186/1471-2105-7-322. [DOI] [PMC free article] [PubMed] [Google Scholar]