Abstract
Functional partnerships between proteins are at the core of complex cellular phenotypes, and the networks formed by interacting proteins provide researchers with crucial scaffolds for modeling, data reduction and annotation. STRING is a database and web resource dedicated to protein–protein interactions, including both physical and functional interactions. It weights and integrates information from numerous sources, including experimental repositories, computational prediction methods and public text collections, thus acting as a meta-database that maps all interaction evidence onto a common set of genomes and proteins. The most important new developments in STRING 8 over previous releases include a URL-based programming interface, which can be used to query STRING from other resources, improved interaction prediction via genomic neighborhood in prokaryotes, and the inclusion of protein structures. Version 8.0 of STRING covers about 2.5 million proteins from 630 organisms, providing the most comprehensive view on protein–protein interactions currently available. STRING can be reached at http://string-db.org/.
INTRODUCTION
In contrast to genome sequences, which are quickly becoming a commodity, the functional connectivity within a proteome is a much more challenging problem. The various protein complexes, transient interactions and functional pathways are all context-dependent, and the experimental techniques for their elucidation are diverse, often not directly comparable, and less reliable than genome sequencing. Nevertheless, protein–protein interaction networks (or also ‘association networks’ in case functional associations are included) are a crucial ingredient for any system-level understanding of cellular machineries (1–5). Furthermore, protein networks can serve very concrete, practical purposes such as filtering and assessing high-throughput functional genomics data, and providing intuitive visual scaffolds for annotating the structural, functional and evolutionary properties of proteins.
The database and web-tool STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) is a meta-resource that aggregates most of the available information on protein–protein associations, scores and weights it, and augments it with predicted interactions, as well as with the results of automatic literature-mining searches. Since its first release in 2000 (6), it has grown into the most comprehensive resource of its type. It builds upon and extends the excellent, manual annotation efforts undertaken at primary protein interaction databases (7–12) and at databases of curated pathway knowledge (13–15). Here, we describe new features that have been added since our report on the previous release, STRING 7 (16).
EXTENDING THE SOURCES OF INTERACTION INFORMATION
The basic interaction unit in STRING is the ‘functional association’, which is defined in this database as the specific and meaningful interaction between two proteins that jointly contribute to the same functional process. With respect to the interacting proteins, STRING does not consider any specific splicing isoforms or posttranslational modifications, but instead represents each protein-coding locus in a genome by a single protein (the longest isoform). Thus, and because STRING aggregates data and predictions stemming from a wide spectrum of cell types and environmental conditions, it aims to represent the union of all possible protein–protein links. From this union, the actual network for any given spatio-temporal snapshot of the cell can in principle be deduced by projection, for example by removing proteins known to be not expressed or not active under the conditions studied (17).
In keeping with the above definitions, STRING imports protein association knowledge not only from databases of physical interactions, but also from databases of curated biological pathway knowledge. Apart form the resources already included in the previous release [MINT (10), HPRD (9), BIND (12), DIP (11), BioGRID (8), KEGG (13) and Reactome (14)], a number of resources have been newly included [IntAct (7), EcoCyc (15), NCI-Nature Pathway Interaction Database and Gene Ontology (GO) protein complexes]. For the full STRING release, this set of previously known and well-described interactions is then complemented by interactions that are predicted computationally, specifically for STRING, using a number of prediction algorithms (18,19). First, we conduct systematic searches for genes that are found in close proximity within prokaryotic chromosomes, which is a good indicator for functional linkage. Second, we search for instances where genes have joined to encode a single fusion protein, which is indicative of functional linkage even in organisms where the two proteins have not fused. Third, we search for gene families that share above-random similarities in their evolutionary histories (i.e. they have similar ‘phylogenetic profiles’). This, again, predicts that they contribute to similar functional processes in the cell. Fourth, we conduct searches for genes that display a similar transcriptional response across a variety of conditions (co-expression). Individually, the above predictors may not always have the specificity of direct experimental interaction assays; however, when used in concert and integrated probabilistically, the performance even of relatively weak predictors can rival that of experimental data (20).
Lastly, two further sources of interactions in STRING are actually providing the majority of associations; these are text-mining and interaction transfer between organisms. For the former, we parse a large body of scientific texts [SGD (21), OMIM (22), The Interactive Fly, and all abstracts from PubMed]. We search for statistically relevant co-occurrences of gene names, and also extract a subset of semantically specified interactions using Natural Language Processing (23). For the transfer of interactions between organisms, we estimate whether a pair of interacting proteins found conserved in another organism justifies the transfer of the interaction to that other organism (24). The transferred interactions, as well as all predicted or imported interactions, are benchmarked and scored against a common reference of functional partnership [we currently use the joint membership of proteins in biological pathways, as annotated at KEGG (13), as our gold-standard].
Together, the above sources of interactions, including predictions and transfers, result in a uniquely high coverage of the interaction networks stored in STRING (Figure 1), particularly for well-studied model organisms. Since the previous release, STRING has almost doubled the number of supported organisms, which now stands at 630. The number of stored interactions has increased as well, to a total of more than 50 million. Since the various subtypes of the interaction evidence are stored separately in the database, they can be disabled at will—giving users the ability to adjust the scope and specificity of STRING towards their particular application.
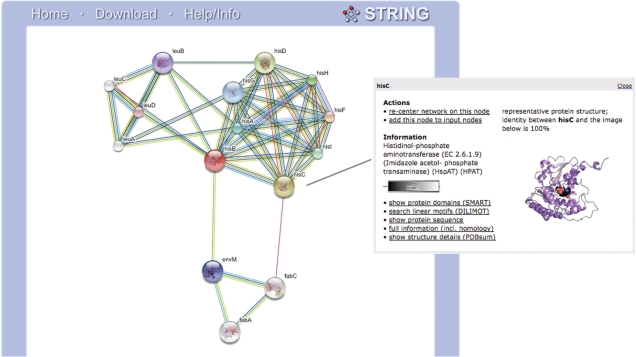
Figure 1.
Protein association network in STRING. An example of the network view in STRING, centered on the query protein ‘hisB’ from Escherichia coli. The inset shows the annotations and options that are available for each protein, including references to other databases. Three ‘functional modules’ can readily be seen in the network, forming tightly connected clusters. These encompass histidine biosynthesis, branched-chain amino acid biosynthesis, and—less strongly connected—a part of fatty acid biosynthesis. Line color indicates the type of the supporting evidence; all underlying evidence can be inspected in dedicated viewers that are accessible from the network.
EXTENDED DEFINITION OF CONSERVED GENOMIC NEIGHBORHOOD
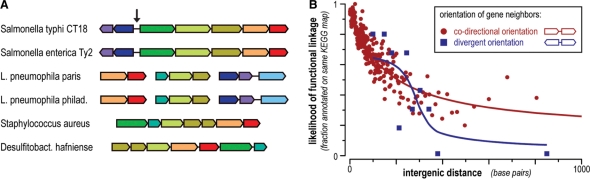
When working with prokaryotes, scientists have long used conserved genomic neighborhood arrangements of genes to infer functional linkage, assuming that such arrangements reflect polycistronic transcription units (operons). STRING has followed this principle, compiling and benchmarking protein–protein associations based on close, co-directional neighborhood of genes on the genome. As of version 8, this has been extended to cover also neighboring genes that are counter-directional in a head-to-head orientation (‘divergent transcription’). Such divergently oriented gene pairs have been shown to be indicative of functional linkage as well (25), albeit with somewhat lower confidence. Often, one of the two genes is a transcriptional regulator, targeting the neighboring gene (25). STRING now uses this type of arrangement in its neighborhood algorithm as well (benchmarked separately, Figure 2). In addition, STRING is now more error tolerant when assembling conserved neighborhoods, ignoring short, partially overlapping genes on the antisense strand that are likely to be spurious predictions.
Figure 2.
Extended definition of genomic neighborhood. (A) Illustration of a conserved gene neighborhood, containing genes related to the biosynthesis and consumption of tryptophan (simplified from a STRING screenshot). Genes connected by lines are direct neighbors on the chromosome, and genes with similar colors are orthologs across the various organisms. The arrow marks a switch in gene orientation, leading to a head-to-head orientation of two presumptive operons. (B) Divergently oriented genes predict functional linkage in prokaryotes. Each dot summarizes a group (bin) of gene pairs with similar intergenic distances. The fraction of such pairs where both genes are annotated in the same KEGG pathway is indicated, implying functional partnership. Note that divergent gene pairs are slightly shifted towards larger intergenic distances, presumably to accommodate promoters and regulatory sequences.
INTEGRATION OF PROTEIN STRUCTURES
For each update, STRING now parses all entries of the PDB database of protein structures (26). The use of protein structures is two-fold: first, to inform the user that a given protein—or a close homolog thereof—indeed has 3D structure information. In this case, a small preview of a representative structure is shown in the network, and the user can follow it to view the full structure and to proceed to the PDB website. Second, protein structures serve as interaction evidence themselves, when more than one distinct peptide chain is found in the structure. In this case, a stable and reliable protein–protein interaction is assumed.
NEW PROGRAMMING INTERFACE
To facilitate the integration of STRING into network tools like Cytoscape (27) and workflow engines like Taverna (28), we have created an application programming interface (API) that allows access to the interaction network in computer-readable formats (Figure 3). Additionally, specific API functions allow retrieval of individual records from our database, for example to map a protein via its name onto a STRING entry. We further envision that the STRING API will be useful to developers of web services, who plan to make use of the STRING interaction network. If a particular web service needs access to the complete set of interactions, it may still be advisable to maintain a local copy of our data distribution. However, if the service requires access to many different subsets (depending on user input), querying STRING via its API could reduce administrative load.
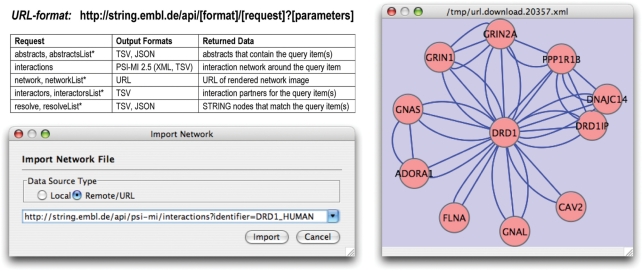
Figure 3.
The new Application Programming Interface, and how it connects to Cytoscape. Specific items of interest can be retrieved from STRING by constructing URLs accordingly (see Table). Unless STRING's internal identifiers are known, an initial call with the ‘resolve’-request is recommended, to map query items to nodes in the STRING network. TSV, tab-separated values; JSON, JavaScript Object Notation; PSI-MI 2.5, Proteomics Standards Initiative Molecular Interaction (XML and tab-delimited format). *Requests ending on ‘List’ accept more than one input item, but are otherwise identical (multiple query items must be separated by URL-encoded ‘new-line’ characters).
The API is called by constructing a URL that contains the type of the request, the desired output format and the input items. The STRING server then returns the result of the computation in the desired format. Further documentation can be accessed via the STRING homepage.
USE SCENARIOS
Apart from the ad hoc and barrier-free access through the website, STRING can be downloaded and used locally, either in the form of concise flat-files or as a mirror installation of the complete relational database back-end (some of the downloads do require a free, nonredistribution license applicable to academic nonprofit users). The interacting entities in STRING can be set to be either proteins, or groups of orthologs spanning multiple organisms (‘COG-mode’). For the latter, STRING relies on an updated and extended version of the COGs [‘Clusters of Orthologous Groups’ (29)], which is being maintained at the eggNOG database (30). A variety of other databases use STRING networks as a basis for further computations/annotations, for example by augmenting the networks with small molecules [STITCH, (31)], or by using the network to increase the power of kinase–substrate predictions [NetworKIN, (32)]. STRING has also been integrated into third-party tools such as NeAT [Network Analysis Tools, (33)], which provides various ways to analyze the interaction network, or Gaggle (34), which enables automated data transfer into other tools via a browser add-on.
FUNDING
Swiss Institute of Bioinformatics; University of Zurich through its Research Priority Program ‘Systems Biology and Functional Genomics’; European Commission's FP6 Programme through the ADIT Integrated Project (LSHB-CT-2005-511065); BioSapiens Network of Excellence (LSHG-CT-2003-503265). Funding for open access charge: University of Zurich.
ACKNOWLEDGEMENTS
The authors wish to thank Dianna Fisk from the Saccharomyces Genome Database, and Thomas B. Brody from The Interactive Fly, for access to gene summary paragraphs. Code development was partially conducted at the ‘WebService BioHackathon 2008’ in Tokyo, Japan.
REFERENCES
- 1.Bader S, Kuhner S, Gavin AC. Interaction networks for systems biology. FEBS Lett. 2008;582:1220–1224. doi: 10.1016/j.febslet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Devos D, Russell RB. A more complete, complexed and structured interactome. Curr. Opin. Struct. Biol. 2007;17:370–377. doi: 10.1016/j.sbi.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Hu Z, Mellor J, Wu J, Kanehisa M, Stuart JM, DeLisi C. Towards zoomable multidimensional maps of the cell. Nat. Biotechnol. 2007;25:547–554. doi: 10.1038/nbt1304. [DOI] [PubMed] [Google Scholar]
- 4.Christensen C, Thakar J, Albert R. Systems-level insights into cellular regulation: inferring, analysing, and modelling intracellular networks. IET Syst. Biol. 2007;1:61–77. doi: 10.1049/iet-syb:20060071. [DOI] [PubMed] [Google Scholar]
- 5.Kohler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lichtenberg U, Jensen LJ, Brunak S, Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- 18.Skrabanek L, Saini HK, Bader GD, Enright AJ. Computational prediction of protein-protein interactions. Mol. Biotechnol. 2008;38:1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- 19.Harrington ED, Jensen LJ, Bork P. Predicting biological networks from genomic data. FEBS Lett. 2008;582:1251–1258. doi: 10.1016/j.febslet.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 20.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 21.Nash R, Weng S, Hitz B, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, et al. Expanded protein information at SGD: new pages and proteome browser. Nucleic Acids Res. 2007;35:D468–D471. doi: 10.1093/nar/gkl931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKusick VA. Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders. 12th. Baltimore: Johns Hopkins University Press; 1998. [Google Scholar]
- 23.Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22:645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- 24.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat. Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 26.Westbrook J, Feng Z, Jain S, Bhat TN, Thanki N, Ravichandran V, Gilliland GL, Bluhm W, Weissig H, Greer DS, et al. The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002;30:245–248. doi: 10.1093/nar/30.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 29.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brohee S, Faust K, Lima-Mendez G, Sand O, Janky R, Vanderstocken G, Deville Y, van Helden J. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 2008;36:W444–W451. doi: 10.1093/nar/gkn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006; 7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]