Abstract
In the scientific research community, plagiarism and covert multiple publications of the same data are considered unacceptable because they undermine the public confidence in the scientific integrity. Yet, little has been done to help authors and editors to identify highly similar citations, which sometimes may represent cases of unethical duplication. For this reason, we have made available Déjà vu, a publicly available database of highly similar Medline citations identified by the text similarity search engine eTBLAST. Following manual verification, highly similar citation pairs are classified into various categories ranging from duplicates with different authors to sanctioned duplicates. Déjà vu records also contain user-provided commentary and supporting information to substantiate each document's categorization. Déjà vu and eTBLAST are available to authors, editors, reviewers, ethicists and sociologists to study, intercept, annotate and deter questionable publication practices. These tools are part of a sustained effort to enhance the quality of Medline as ‘the’ biomedical corpus. The Déjà vu database is freely accessible at http://spore.swmed.edu/dejavu. The tool eTBLAST is also freely available at http://etblast.org.
INTRODUCTION
Authorship of scientific papers is one of the most valuable currencies for scientists and engineers, and is an asset not only for climbing the corporate or academic ladder (1), but also most importantly to secure funding for academic laboratories. The fierce competition in most scientific disciplines and the increasing necessity to publish may lead authors to engage in questionable behavior such as publishing a single piece of work more than once, or emulating the style, or copying the content of another person's work. Duplicate publication may be useful to provide wider access to the scientific community or to report important updates to surveys or clinical trials, but publications that simply reproduce a previous work with virtually identical results and conclusions often lack the novelty to justify additional publication. The latter types of duplicate publication are considered unethical because they undermine the public confidence in scientific integrity. Others have previously described additional duplicate publication behaviors referred to as ‘salami slicing’ (dissecting a scientific work into multiple least publishable units) and ‘meat extenders’ (building on a previous publication with new data that would not be publishable alone) (2–4). Most previous studies of duplicate publication have been limited to a particular scientific field where duplication was painstakingly identified manually, underscoring the need for an automated method to detect putative duplications (5–16).
We have established a method to identify highly similar citations in Medline, the comprehensive literature database of life sciences and biomedical information, using the text similarity search engine eTBLAST (17,18). We were able to statistically calibrate eTBLAST to identify citations that have unusually high similarity, which were then saved in Déjà vu pending manual inspection (19,20).
CONTENT AND METHODS
Identification of highly similar citations
Technical details describing the detection of highly similar citations and its application to the entire Medline database have been reported previously (19,20). Briefly, the method which has contributed the preponderance of entries in Déjà vu involves ‘eTBLASTing’ each Medline citation against its most related article (a feature available from Medline). Upon comparison, citation pairs are so highly similar that predetermined similarity thresholds exceeded are flagged as a highly similar pair and stored in Déjà vu awaiting manual verification by human curators.
Manual classification of highly similar citations
Déjà vu was designed and developed to allow for collaborative work among the multiple curators. It was also necessary to define a broad, flexible and extensible classification scheme to accommodate a wide range of highly similar documents dealing with all areas of biomedical research, reflecting different publication behaviors, styles and agreements. Upon manual verification, highly similar citation pairs were classified in one or more of the categories listed and defined in Table 1. In particular, we sought to distinguish between appropriate and inappropriate duplication, a process which is admittedly subjective. A pair of duplicates with different authors may indicate potential plagiarism, while two publications with shared authors may indicate multiple publication of the same study. Updates to clinical trials or survey type research are instances where complete duplication is not necessarily inappropriate. Similarly, studies with different outcomes using similar phraseology may bring valuable new information. Errata, which may or may not be tagged as such in Medline, are most similar to the initial record, often involving only a typographical correction. All of these determinations are difficult or impossible to accomplish computationally, and thus are best made by human curators.
Table 1.
Déjà vu content by category and category definitions
| Duplication type | Count | Description |
|---|---|---|
| DISTINCT | 1379 | There are a number of reasons for different citations to have a high similarity, including citations that describe related, but very distinct publications. A pair of citations identified by computer similarity, which after inspection is, for example, clearly a continuation of a study which has evolved, and the text represents new information that is categorized as a distinct and unique work |
| DUPLICATE | 2443 | A pair of citations that was identical or nearly identical. The citations report on a study with the same or very similar results and conclusions. |
| ERRATUM | 188 | Only a fraction of the MEDLINE records that are apparently corrections to previous entries are marked as errata. If a title/abstract pair is either labeled as errata or if it is clear that a correction has been made (author list, spelling, small changes to abstract or title wording, etc.), then the errata classification is used. |
| SANCTIONED | 1619 | There are a number of reasons for different citations to have a high level of similarity, some of which play a special, very important, and very legitimate role in the reporting of science. Examples include periodic reviews, periodic guidelines, specialized databases and specialized federal register citations. Citation pairs of this type, identified through computer text similarity have been manually classified to the category sanctioned. |
| NO ABSTRACT | 16 | In some cases highly similar titles are flagged as potential duplicates, but the non-identity MEDLINE record does not contain an abstract, we designate that pair as a ‘NO ABSTRACT’ to indicate that its status cannot be determined. |
| UNVERIFIED | 69115 | Deja vu is a database of duplicate publications, as identified using a number of different techniques, with the principle one being text similarity comparisons. Those putative duplicates identified by any of these techniques, prior to human verification and assignment to another category, are initially loaded into these categories, and since our software also inspects the author lists, they are loaded into unverified categories that have either overlapping authors (SA) or not (DA). |
| TOTAL | 74 760 |
Up to date statistics and definitions are available at http://spore.swmed.edu/dejavu/help and http://spore.swmed.edu/dejavu/statistics/.
Déjà vu in numbers
All data collected have been consolidated into a web-accessible database, available at http://spore.swmed.edu/dejavu. As of 22 July 2008, Déjà vu contains a total 74 760 records of which 5645 have been manually inspected (Table 1). Déjà vu has received over 40 000 visits since 1 January 2008 and currently receives an average of about 2000 visits per month.
QUERIES AND INTERFACE
The Déjà vu interface was designed using python (http://python.org) and the Django web framework (http://djangoproject.com). Data are stored in a backend MySQL Database (http://mysql.com). Déjà vu was designed to allow real-time collaborative annotation by multiple curators who need not be programmers to add comments and updates or create new records.
On the Déjà vu website users can: (i) browse Déjà vu entries with no specific search method (Each entry links to the scientific citation along with full text when freely available.); (ii) perform generic searches within the Déjà vu content by authors, address, title word, abstract word, year and comment word; (iii) perform detailed searches by specifying search criteria specific to PMID, journal names, title words, abstract, address and year; (iv) filter and view Déjà vu results in a particular category or identified by particular authors (same or different), language, availability of full text, discovery method, etc.; (v) send comments or reports to contest a record or submit a potential duplication to be reviewed by human curators; and (vi) access statistics using different filters including category, language, country, journals, etc.
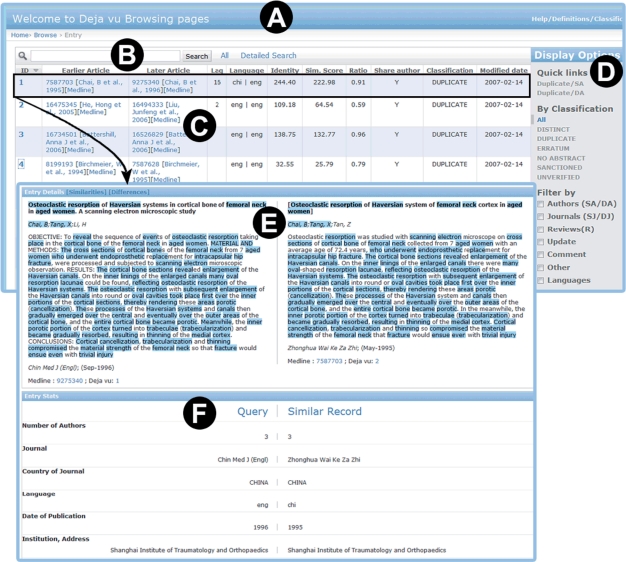
For each duplicate record, a viewing window presents citations side-by-side with similarities or differences highlighted (Figure 1), providing a user-friendly interface to search, browse and facilitate rapid and rigorous interpretation of the results. Déjà vu data are also available for data mining in two formats: comma-separated values and a MySQL script to recreate the MySQL database.
Figure 1.
The Déjà vu citation presentation output. (A) Browsing interface for database content. (B) Query box to search duplicate records by author names, title, abstract, year of publication and comment words. (C) List of records in Déjà vu including PMIDs, author names, publication date and links to Medline citations and free full text when available. (D) Category filters to browse records in a particular category. (E) Side-by-side view of a duplicate record highlighting overlapping keywords in blue. (F) Miscellaneous information for each article involved.
CONCLUSION AND FUTURE DIRECTIONS
The Déjà vu database is the first of its kind to publically present cases of highly similar citations in Medline. In addition to presenting the list of highly similar citations, a goal of Déjà vu is to help scientists study in depth the behaviors of authors and the characteristics underlying multiple publications and related ethics issues surrounding the process of scientific publication. A friendly interface provides users with various browsing options along with a graphical representation of the overlapping information between citations. Ultimately, Déjà vu may act as a deterrent to the unethical practice of duplication.
Further work, currently in progress, that will substantially improve Déjà vu includes: (i) a streamlined process to update Déjà vu on a daily basis. (ii) a more collaborative approach for recruitment and qualification of topical experts as volunteer curators for specific publication areas. (iii) New methods to better address the question most often asked by authors introduced to Déjà vu, ‘Am I in it, or has my work been duplicated? ’ Authors can now check if their work has been duplicated by submitting their abstracts one by one directly to eTBLAST, which then flags highly similar citations for the authors to pursue. Utilities are being developed to allow authors to scan their entire bibliography at once (retrieved using Medline Entrez keyword queries) to obtain a list of highly similar citations for each citation entered. Authors will also be able to automatically submit suspicious highly similar citations found by this process directly to Déjà vu curators. (iv) Currently, duplications found in Déjà vu were obtained from Medline citations. Other literature databases will be added as they are scanned by eTBLAST, including the Institute of Physics, NASA and NIH CRISP.
FUNDING
P.O’B. Montgomery Distinguished Chair (to H.G.); the Hudson Foundation (to H.G.); National Institute of Health/National Library of Medicine grant (R01 LM009758-01 to H.R.G.). Funding for open access charge: P.O'B;. Montgomery Distinguished Chair.
ACKNOWLEDGEMENTS
The authors thank David Trusty for computer administrative support, Dr John Loadsman as a substantial contributing curator, Dr Wayne Fisher for useful comments and discussions and Linda Gunn for administrative assistance. They also wish to thank numerous Déjà vu users who have reported inaccuracies or have alerted them to questionable publications.
REFERENCES
- 1.Budinger TF, Budinger MD. Ethics of Emerging Technologies, Scientific Facts and Moral Challenges. NJ: John Wiley and Sons; 2006. [Google Scholar]
- 2.Broad WJ. The publishing game: getting more for less. Science. 1981;211:1137–1139. doi: 10.1126/science.7008199. [DOI] [PubMed] [Google Scholar]
- 3.Huth EJ. Irresponsible authorship and wasteful publication. Ann. Intern. Med. 1986;104:257–259. doi: 10.7326/0003-4819-104-2-257. [DOI] [PubMed] [Google Scholar]
- 4.von Elm E, Poglia G, Walder B, Tramer MR. Different patterns of duplicate publication: an analysis of articles used in systematic reviews. J. Am. Med. Assoc. 2004;291:974–980. doi: 10.1001/jama.291.8.974. [DOI] [PubMed] [Google Scholar]
- 5.Schein M, Paladugu R. Redundant surgical publications: tip of the iceberg? Surgery. 2001;129:655–661. doi: 10.1067/msy.2001.114549. [DOI] [PubMed] [Google Scholar]
- 6.Rosenthal EL, Masdon JL, Buckman C, Hawn M. Duplicate publications in the otolaryngology literature. Laryngoscope. 2003;113:772–774. doi: 10.1097/00005537-200305000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Roig M. Re-using text from one's own previously published papers: an exploratory study of potential self-plagiarism. Psychol. Rep. 2005;97:43–49. doi: 10.2466/pr0.97.1.43-49. [DOI] [PubMed] [Google Scholar]
- 8.Mojon-Azzi SM, Jiang X, Wagner U, Mojon DS. Redundant publications in scientific ophthalmologic journals: the tip of the iceberg? Ophthalmology. 2004;111:863–866. doi: 10.1016/j.ophtha.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 9.Kostoff RN, Johnson D, Rio JAD, Bloomfield LA, Shlesinger MF, Malpohl G, Cortes HD. Duplicate publication and ‘paper inflation’ in the Fractals literature. Sci. Eng. Ethics. 2006;12:543–554. doi: 10.1007/s11948-006-0052-5. [DOI] [PubMed] [Google Scholar]
- 10.Gotzsche PC. Multiple publication of reports of drug trials. Eur. J. Clin. Pharmacol. 1989;36:429–432. doi: 10.1007/BF00558064. [DOI] [PubMed] [Google Scholar]
- 11.Durani P. Duplicate publications: redundancy in plastic surgery literature. J. Plast. Reconstr. Aesthet. Surg. 2006;59:975–977. doi: 10.1016/j.bjps.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 12.Chennagiri RJR, Critchley P, Giele H. Duplicate publication in the Journal of Hand Surgery. J. Hand Surg. 2004;29:625–628. doi: 10.1016/j.jhsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Bloemenkamp DG, Walvoort HC, Hart W, Overbeke AJ. [Duplicate publication of articles in the Dutch Journal of Medicine in 1996] Ned. Tijdschr. Geneeskd. 1999;143:2150–2153. [PubMed] [Google Scholar]
- 14.Blancett SS, Flanagin A, Young RK. Duplicate publication in the nursing literature. Image J. Nurs. Sch. 1995;27:51–56. doi: 10.1111/j.1547-5069.1995.tb00813.x. [DOI] [PubMed] [Google Scholar]
- 15.Barnard H, Overbeke AJ. [Duplicate publication of original manuscripts in and from the Nederlands Tijdschrift voor Geneeskunde] Ned. Tijdschr. Geneeskd. 1993;137:593–597. [PubMed] [Google Scholar]
- 16.Bailey BJ. Duplicate publication in the field of otolaryngology-head and neck surgery. Otolaryngol. Head Neck Surg. 2002;126:211–216. doi: 10.1067/mhn.2002.122698. [DOI] [PubMed] [Google Scholar]
- 17.Lewis J, Ossowski S, Hicks J, Errami M, Garner HR. Text similarity: an alternative way to search MEDLINE. Bioinformatics. 2006;22:2298–2304. doi: 10.1093/bioinformatics/btl388. [DOI] [PubMed] [Google Scholar]
- 18.Errami M, Wren JD, Hicks JM, Garner HR. eTBLAST: a web server to identify expert reviewers, appropriate journals and similar publications. Nucleic Acids Res. 2007;35:W12–W15. doi: 10.1093/nar/gkm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Errami M, Garner H. A tale of two citations. Nature. 2008;451:397–399. doi: 10.1038/451397a. [DOI] [PubMed] [Google Scholar]
- 20.Errami M, Hicks JM, Fisher W, Trusty D, Wren JD, Long TC, Garner HR. Deja vu–a study of duplicate citations in Medline. Bioinformatics. 2008;24:243–249. doi: 10.1093/bioinformatics/btm574. [DOI] [PubMed] [Google Scholar]