Abstract
GermSAGE is a comprehensive web-based database generated by Serial Analysis of Gene Expression (SAGE) representing major stages in mouse male germ cell development, with 150 000 sequence tags in each SAGE library. A total of 452 095 tags derived from type A spermatogonia (Spga), pachytene spermatocytes (Spcy) and round spermatids (Sptd) were included. GermSAGE provides web-based tools for browsing, comparing and searching male germ cell transcriptome data at different stages with customizable searching parameters. The data can be visualized in a tabulated format or further analyzed by aligning with various annotations available in the UCSC genome browser. This flexible platform will be useful for gaining better understanding of the genetic networks that regulate spermatogonial cell renewal and differentiation, and will allow novel gene discovery. GermSAGE is freely available at http://germsage.nichd.nih.gov/
INTRODUCTION
Spermatogenesis is a complex and tightly regulated developmental process that involves mitotic expansion and proliferation of spermatogonia (Spga), meiosis in spermatocytes (Spcy) and postmeiotic differentiation in spermatids (Sptd). It provides an informative model system for studying the underlying molecular mechanisms of the physiological changes occurring during self-renewal and differentiation of germ cell development. Despite its biological importance, little is known about stage-specific gene regulation in spermatogenesis. The information on male germ cell specific transcripts is very limited, with only 1962 transcripts in type A Spga, 4385 in pachytene Spcy and 4014 in round Sptd in the Unigene database (1). To provide a comprehensive understanding of male germ cell development, we profiled and analyzed the transcriptome of male germ cells at Spga, Spcy and Sptd stages by Serial Analysis of Gene Expression (SAGE) (2–5). SAGE provides several advantages over other gene expression profiling methods including microarray analysis. It is a high-throughput method, which simultaneously detects and measures the expression level of genes including rare genes, in a cell at a given time. It does not depend on the prior availability of transcript information (6) and provides an unbiased method to examine both known and unknown genes. We identified a wide variety of genes specifically expressed during male germ cell development (Table 1). Importantly, we identified novel regulatory and expression patterns such as an extensive presence of antisense transcripts and spliced variants. These antisense transcripts had multiple origins, including processed sense transcripts, intronic and exonic sequences of a single gene or multiple genes, intergenic sequences and pseudogenes. One-third of the identified genes demonstrated evidence of alternative splicing (3,4).
Table 1.
Statistics of germ cell SAGE libraries
| SAGE tag | Spga | Spcy | Sptd |
|---|---|---|---|
| Reliable tags | 110 872 | 11 384 | 11 214 |
| Total unique tags | 31 514 | 36 147 | 34 000 |
| Known genes | 12 508 | 13 691 | 12 368 |
| Uncharacterized cDNA | 3020 | 4406 | 4234 |
| Novel | 563 | 1058 | 1155 |
| Transcripts identified | 16 091 | 19 155 | 17 757 |
Reliable tags refers to SAGE tags without ambiguous nucleotide base. Known genes are defined as those that match Unigene clusters with at least one known mRNA; uncharacterized cDNA refers to the full cDNAs from RIKEN, IMAGE, expressed clones or any cDNAs without annotation. Novel tag refers to no Unigene cluster match. Each transcript is covered with an average of 3.6 SAGE tags.
The GermSAGE database is a comprehensive web-based database containing male germ cell transcript data. The tag coverage for each germ cell SAGE library is over 150 000 and contains a total of 452 095 SAGE tags derived from type A Spga, pachytene Spcy and round Sptd. It is a flexible platform which is integrated with the UCSC genome browser allowing browsing, search and comparative analysis of male germ cell data with customizable search parameters. GermSAGE leads to a better understanding of male germ cell development and may provide insight on male infertility, and allows development of genetic tests and therapies.
DESIGN AND IMPLEMENTATION
Displaying features
The GermSAGE application provides an organized approach for sharing genomic data in the form of a Browser Extensible Display (BED) format. It utilizes the UCSC Genome Browser (7) to visualize experimental datasets with respect to the mouse genome (NCBI Build 35 assembly). Genome coordinate information in the BED file was obtained by BLASTN analysis of the SAGE tag sequences (8) against the mouse genome (NCBI Build 35 assembly); only perfectly matched tags were retained. In case a tag match to more than one Unigene clusters, the complete Unigene list can be retrieved by using ‘Full text or specific field search’ in the main page. Users can compare the GermSAGE data to other genome annotations or upload their own data using UCSC Genome Browser's custom annotation track feature. The custom annotation track is viewable on top of the GermSAGE dataset, thus making it an ideal mechanism for displaying and analyzing personalized data.
Architecture and technologies
GermSAGE is a web based application constructed using components and services that work together to form a multi-layered architecture. The core of the system is based on a domain model that is comprised of Java objects that describe the business, operations and object relationships (9). The domain model is established using Hibernate (10). This framework organizes mapping of the domain model to the underlying relational database of the application. The result is a transparent back-end layer that facilitates data query and retrieval features. The view layer of the application is rendered primarily using Java Server Pages (JSP, v2.0). This layer is XHTML 1.0 strict compatible. Client-side functionality is coded using JavaScript. The application layers are joined using Spring (11), a highly customizable and extensible framework which provides common infrastructures. The system will check the availability of the host site (http://genome.ucsc.edu) before rendering the BED files.
USING GERMSAGE
The entry page of the GermSAGE database provides two analysis strategies: (i) a global view of germ cell transcriptome data by selecting ‘Genome view of complete data set’; and (ii) search genes or specify searching criteria based on tag sequence, chromosomal location or tag counts via the tab ‘Full text or specific field search’. All results generated from both approaches will be displayed in UCSC, a tutorial on how to use the UCSC genome browser is available (12,13).
Genome view of complete data set
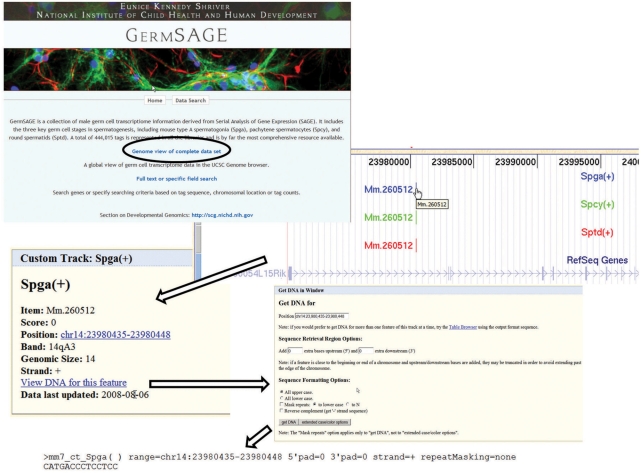
This is the ‘free view’ mode of GermSAGE that will open a new browser window that directs the users to the UCSC genome browser; this provides both graphical and text-based views of the SAGE tag data and annotations (Figure 1). Users can freely navigate in the genome browser by using the display navigation toolbar on the top to examine the overall expression patterns of transcripts. The position/search box allows users to locate the expression of germ cell transcripts at a particular location or in association with the keyword found in the genome browser database. SAGE tag sequences at different germ cell stages are aligned in the genome browser and presented with different colors (Spga: blue; Spcy: green; Sptd: red). The SAGE tags are also marked with + or − on the track to indicate the strand orientation, and can be turned on or off by changing the custom tracks setting. The Unigene assignment can be retrieved by clicking on the color tag or select ‘pack’ option in the custom tracks item below the annotation window. For more details about the tag and its sequence, click on the tag again to bring up a window that shows the tag information, including Unigene assignment, chromosomal position/band and strand orientation. Click on ‘view DNA for this feature’ will bring up ‘Get DNA in Window’, which retrieves the tag sequence. A number of manipulations on the tag sequence are available before exporting to the web browser in FASTA format (Figure 1).
Figure 1.
Genome view in GermSAGE. The genome view allows free navigation on male germ cell transcriptome data in UCSC genome browser. SAGE tags for Spga, Spcy and Sptd are marked in blue, green and red, respectively. SAGE tag tracks can be turn on and off through the custom track option. The sequences on how to retrieve the tag sequence are shown. Users can modify and process the sequence before exporting in FASTA format.
One of the powerful features of GermSAGE is that it provides users with an interaction interface in genome browser format. The user can overlay the male germ cell transcriptome data with a variety of annotated tracks below the genome view window, and create a custom map by adding tracks to view various types of data and specific genomic landmarks. The browser offers a broad list of options. It includes: (1) mapping and sequencing tracks that contains information about the position, marker and GC percentage of the annotated gene region; (2) genes and gene prediction tracks on gene annotation and predictions from various sources; (3) mRNA and EST tracks that contains information on transcripts, CAGE (Cap Analysis of Gene Expression) tags to identify potential transcription start sites and alternatively spliced RNA species; (4) expression and its regulation that includes expression data from different microarray platforms and regulatory information such as CpG islands and microRNA; (5) comparative genomics shows the sequence conservation among species; and (6) variation and repeats.
This information will lead to better insights about gene regulation, and facilitate the generation of hypotheses.
Full text or specific field search
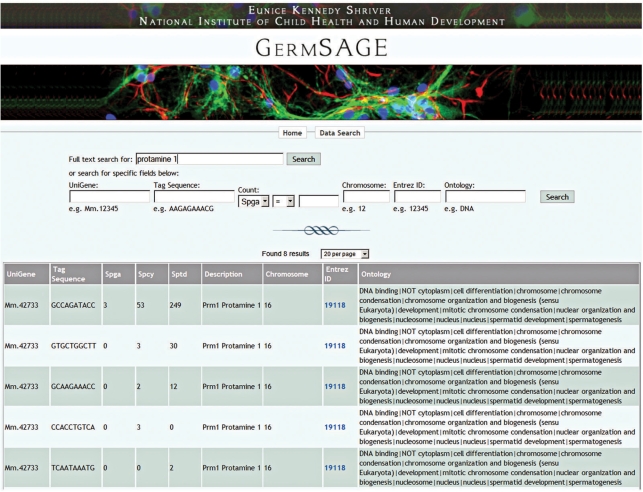
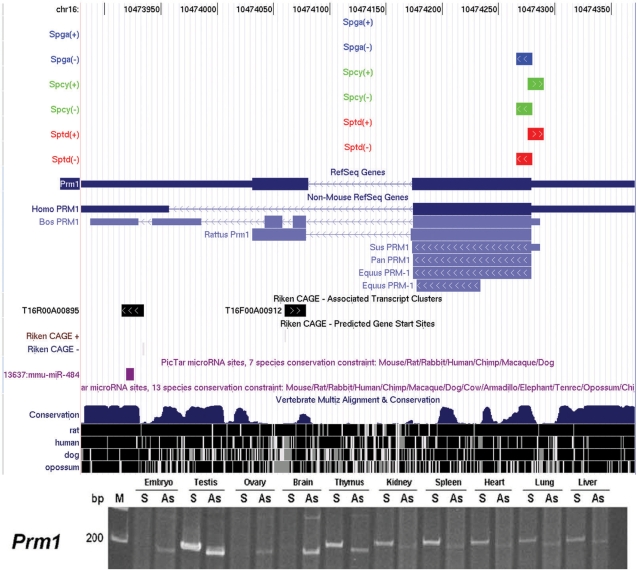
This option provides a more focused and quantitative analysis of male germ cell transcripts based on the inputs defined by the users. Inputs include gene name/symbol, tag sequence, Unigene ID, chromosomal region or gene function in terms of gene ontology. Here, we use Protamine 1 (Prm1) as an example to show the power of GermSAGE in dissecting the complexity of antisense transcription in the male germ trancriptome. Recent study of the human genome suggested 61% of loci expressed antisense transcripts (14). We have demonstrated frequent antisense transcription events in the male germ cell transcriptome could be classified into three groups (4). Protamines are arginine-rich nuclear proteins that replace histones late in the haploid phase of spermatogenesis, and are important for sperm head condensation and DNA stabilization. They are abundantly expressed in spermatid stage (15). There are a total of five SAGE tags mapped to Prm1 and Sptd showed the highest expression (Figure 2), as previously reported (15). A click on the Entrez ID will show the SAGE tags associated with Prm1 in the genome browser. Prm1 is highly conserved among species as seen on the conservation track and might be regulated by microRNA miR-484 (16) at the 3′-end. Three tags are demonstrated to be derived from the sense transcript, while two tags (one in Spcy and one in Sptd) are derived from the antisense transcript. This is also suggested by the presence of the putative transcription start site in the antisense CAGE tag cluster (T16F00A00912) (Figure 3). The existence of both sense and antisense transcripts was confirmed by orientation specific RT–PCR (Figure 3).
Figure 2.
Full text or specific field search in GermSAGE. GermSAGE output for protamine 1 was illustrated. The search fields allow qualitative lookup based on keyword, ID, position and gene ontology. Quantitative analysis based on tag count is also available. A combination of different search criteria is allowed.
Figure 3.
SAGE tags mapped to protamine 1. SAGE tags mapped to Prm1 are overlaid with additional annotation tracks, such as conservation among species, Riken CAGE and microRNA to get better insights on potential gene regulation. The bottom panel shows validation of prm1 sense (S) and antisense transcripts (AS) in different tissues. M indicates the marker.
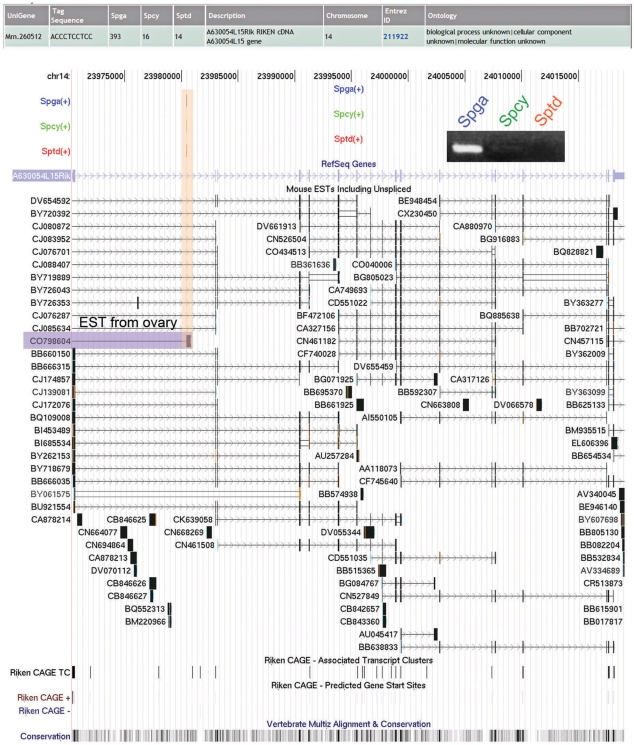
GermSAGE can also be applied for novel gene discovery in male germ cell development. The total number of uncharacterized cDNA and novel transcripts in the GermSAGE database is 20 707 and 2776, respectively. To highlight this capability, we used the search tool to identify a tag preferentially expressed in spermatogonial stage. The expression level is over 24- and 28-fold compared to Spcy and Sptd, respectively (Figure 4). The genome browser showed the tag oriented in the sense direction to the uncharacterized cDNA A630054L15Rik. Importantly, the tag alignment suggested the transcript might be an alternative splice variant of the cDNA. To increase the predictive evidence, the EST track and CAGE tag annotations were turned on to support the potential existence of the transcript. It turned out that the tag mapped perfectly to an EST sequence derived from ovary (CO798604, Figure 4) at the 3′-end and a transcription start site supported by CAGE tag at the 5′-end. RT–PCR confirmed the prediction and it is highly expressed in Spga (Figure 4). The same approach can be applied to other novel gene discovery as well.
Figure 4.
Novel gene search in GermSAGE. A tag preferentially expressed in Spga was identified. The tag indicated the presence of a potential short isoform. This observation was supported by EST and CAGE tag evidence. RT–PCR confirmed the tag was exclusively expressed in Spga.
GermSAGE allows a combination of search parameters. Transcripts may also be retrieved by tag count number. This is useful for refining output based on expression level. For example, one can retrieve Spga expressed transcripts with tag counts more than 10 involved in DNA binding on chromosome 12 by selecting Spga >10, keying in 12 in the chromosome field and DNA binding in the ontology field. The data can be sorted in ascending or descending order by clicking on the items in the first row. This function is also useful in revealing transcripts that demonstrate stage-specific gene expression.
FUTURE DEVELOPMENTS
GermSAGE is created as a flexible platform for male germ cell transciptome analysis. In addition to static data analysis, we previously leveraged the power of SAGE by developing algorithms to extract cellular dynamics contained in the SAGE data to reveal transcriptional regulation and gene interaction networks at particular male germ cell stages and during cellular transition (5). We are working on the pipeline for such dynamic data analysis and this will be included in the next version of GermSAGE. In the current version, the rendering of transcriptome data takes up to 2 min. To address this performance issue, we are currently planning to modify and deploy the genome browser source code on servers at the National Institute of Child Health and Human Development, National Institutes of Health. The long-term scientific vision of GermSAGE is to serve as a centralized platform to scan the entirety of dynamic genomic changes in male germ cell development, as well as the manifestations of those changes through gene expression patterns.
DATABASE AVAILABILITY
The database is freely available and can be accessed from http://germsage.nichd.nih.gov. GermSAGE is compatible with common web browsers in the market, including Microsoft internet explorer, Mozilla firefox and Apple Safari, and is platform independent. All feedbacks are welcome and should be forwarded to Dr Tin-Lap Lee at leetl@mail.nih.gov.
FUNDING
Open access charge and this research are funded by the Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development.
REFERENCES
- 1.Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu SM, Baxendale V, Chen Y, Pang AL, Stitely T, Munson PJ, Leung MY, Ravindranath N, Dym M, Rennert OM, et al. Analysis of mouse germ-cell transcriptome at different stages of spermatogenesis by SAGE: biological significance. Genomics. 2004;84:971–981. doi: 10.1016/j.ygeno.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Chan WY, Lee TL, Wu SM, Ruszczyk L, Alba D, Baxendale V, Rennert OM. Transcriptome analyses of male germ cells with serial analysis of gene expression (SAGE) Mol. Cell Endocrinol. 2006;250:8–19. doi: 10.1016/j.mce.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Chan WY, Wu SM, Ruszczyk L, Law E, Lee TL, Baxendale V, Lap-Yin Pang A, Rennert OM. The complexity of antisense transcription revealed by the study of developing male germ cells. Genomics. 2006;87:681–692. doi: 10.1016/j.ygeno.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Lee TL, Alba D, Baxendale V, Rennert OM, Chan WY. Application of transcriptional and biological network analyses in mouse germ-cell transcriptomes. Genomics. 2006;88:18–33. doi: 10.1016/j.ygeno.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 7.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lash AE, Tolstoshev CM, Wagner L, Schuler GD, Strausberg RL, Riggins GJ, Altschul SF. SAGEmap: a public gene expression resource. Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans E. Domain-driven Design: Tackling Complexity in the Heart of Software. Boston, MA: Addison-Wesley Professional; 2003. [Google Scholar]
- 10.Bauer C, King G. Java Persistence with Hibernate. Greenwich, CT: Manning Publications; 2006. [Google Scholar]
- 11.Walls C, Breidenbach R. Spring in Action. 2nd edn. Greenwich, CT: Manning Publications; 2007. [Google Scholar]
- 12.Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. UCSC genome browser tutorial. Genomics. 2008;92:75–84. doi: 10.1016/j.ygeno.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Bina M. The genome browser at UCSC for locating genes, and much more! Mol Biotechnol. 2008;38:269–275. doi: 10.1007/s12033-007-9019-2. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 15.Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]