Abstract
ConSurf-DB is a repository for evolutionary conservation analysis of the proteins of known structures in the Protein Data Bank (PDB). Sequence homologues of each of the PDB entries were collected and aligned using standard methods. The evolutionary conservation of each amino acid position in the alignment was calculated using the Rate4Site algorithm, implemented in the ConSurf web server. The algorithm takes into account the phylogenetic relations between the aligned proteins and the stochastic nature of the evolutionary process explicitly. Rate4Site assigns a conservation level for each position in the multiple sequence alignment using an empirical Bayesian inference. Visual inspection of the conservation patterns on the 3D structure often enables the identification of key residues that comprise the functionally important regions of the protein. The repository is updated with the latest PDB entries on a monthly basis and will be rebuilt annually. ConSurf-DB is available online at http://consurfdb.tau.ac.il/
INTRODUCTION
The study of a protein raises many questions: What the protein function is? Does it have more than one function? How does the protein perform its functions? Is it acting alone? Where/when is the protein active? Where are the functional regions of the protein and what their nature is? Each of these questions can be further refined into additional, more specific, questions.
Advances in sequencing technologies produce ever larger databases containing protein sequences from a large collection of species. Within these databases one can find many protein families that can be analyzed in search for functional regions. Generally speaking, protein function is mediated through clusters of evolutionarily conserved amino acids that are located in close vicinity to each other. These clusters may be involved in enzymatic activity, ligand binding, protein–protein interactions, or in the folding and stabilization of the protein's architecture (1). Typically, the detection of these clusters is useful for initial investigation of a protein by characterizing their properties. In addition, correlating the conservation pattern with other data is often insightful. The ConSurf-DB leverages the protein databases in order to aid in the detection of such clusters.
We introduced the original ConSurf, available as an online server (2) at http://consurf.tau.ac.il/, back in 2001 (3). ConSurf was developed for the identification of functional regions in proteins based on the conservation of amino acids, taking into account the phylogenetic relations between the proteins. In 2005 we introduced the ConSurf-HSSP (4) database which was a pre-calculated repository of ConSurf results based on multiple sequence alignments (MSAs) extracted from the HSSP database (5). The MSAs in HSSP do not include the gaps in the query sequence, i.e. positions in the aligned sequences which do not have corresponding positions in the query sequence are removed from the alignment. Consequently, the phylogenetic reconstruction of the protein family is prone to errors. The ConSurf-DB, presented here, replaces ConSurf-HSSP as our repository of pre-calculated ConSurf results. The MSAs in the ConSurf-DB include all sequence data needed for the phylogenetic reconstruction, it also uses a more advanced Rate4Site (6) algorithm utilizing Bayesian inference rather than the Maximum Likelihood estimate that was used in ConSurf-HSSP. The conservation results of ConSurf-DB are presented in much more standard and cross platform formats.
Other tools for predicting functional sites based on evolutionary conservation include the Evolutionary Trace Viewer (7) and SiteFiNDER|3D (8). Like ConSurf-DB, they take advantage of the evolutionary relationship between homologues to detect regions that are likely to be of functional importance. Other tools take a different approach: The HotPatch (9) tool predicts functionally important regions by performing a statistical analysis and comparing the protein's surface against the surfaces of a large set of proteins (not necessarily homologous to that protein) whose functional sites are known. For a brief comparison of ConSurf-DB with these tools please see the supporting materials.
The sequence homologues of each protein in ConSurf-DB are collected using PSI-BLAST (10) and then automatically filtered in order to represent reliably and comprehensively the protein family. This process requires a delicate balance between two opposing effects. A conservative search would yield only very close homologues and would make it virtually impossible to discriminate between amino acid positions that are truly important and those that did not change because of insufficient evolutionary time. On the other hand, an overly permissive search may falsely detect non-homologues that do not share the same structure and/or function. We conducted preliminary investigations and came up with a scheme, presented below, which balances between these two extremes. The selected homologues are aligned using MUSCLE (11) and are available for use as part of the ConSurf-DB repository.
The Rate4Site program is subsequently used to construct a phylogenetic tree and calculate conservation scores. Rate4Site builds a phylogenetic tree of the homologues using the neighbor joining algorithm (12). Using an empirical Bayesian approach it calculates the evolutionary rate of each amino acid position of the MSA, taking into account the stochastic nature of the evolutionary process. The amino acid evolution is traced using the JTT (13) substitution model. High evolutionary rate represents a variable position while low rate represents an evolutionarily conserved position.
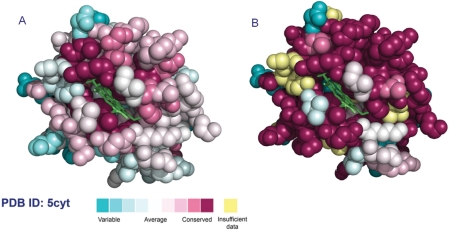
The conservation scores are normalized so that the average over all residues is zero, and the standard deviation is one. Low (negative) scores indicate the conserved positions while the high scores indicate the variable ones. The normalized scores are then binned into the 1–9 color codes presented in Figure 1, representing the conservation grades and projected on the 3D model of the query protein, where 1 corresponds to maximal variability and 9 to maximal conservation. It is important to note that even though the same scale is used in all the protein families, the conservation scores are not absolute and hence, comparing the conservation scores between different protein families might be misleading.
Figure 1.
Cytochrome c. (A) The conservation coloring profile from the ConSurf-DB repository, mapped onto a space-filling representation of the protein. The conservation coloring scale is shown below. The HEME group, in stick representation, is colored green. (B) The same view as calculated by the ConSurf server using default parameters.
There are several ways to access the repository. For visual inspection of one or few proteins, a web interface, available at http://consurfdb.tau.ac.il/, supports 3D visualization (using FirstGlance in Jmol) and access to all supplementary data. The entire repository can be downloaded via ftp or rsync and used for large-scale automated studies. For advanced uses, involving re-building of variants of the repository, the build scripts can be downloaded from the ConSurf-DB web site. We will be glad to assist in adopting them to different environments.
METHODOLOGY
Building the ConSurf-DB repository consists of four stages: scanning the PDB (14), building MSA files, calculating the conservation scores and formatting the results (supporting material, Figure 2). This design was chosen to allow reusability of the scripts by controlling the data at each step. For instance, an MSA file can be created by simply inputting a FASTA format sequence file to the MSA building script or if a Rate4Site output was obtained using a unique set of parameters, it can be used to create 3D visualization. A monthly update process will calculate the conservation profiles for new PDB entries. In the annual refresh, the entire database will be re-calculated based on MSAs created from the latest sequence databases.
The ConSurf-DB build process is completely automated and starts by scanning the PDB. Each PDB entry can contain one or more chains that are handled separately. When a new PDB entry is found, the SEQRES section of each chain passes through three filters: ‘type’, ‘length’ and ‘modifications’. Nucleic acid chains are discarded by the ‘type’ filter, short amino acid chains of less than 30 residues are discarded by the ‘length’ filter as they do not contain enough data for reliable phylogenetic tree reconstruction. Finally, the ‘modification’ filter converts a list of non-standard residues into their closest standard amino acid form, and discards highly modified chains with over 15% non-standard residues. The modifications are noted and saved as part of the chain's supplementary data.
The next two steps rely solely on the sequence of amino acids in the chain. Identical sequences are grouped and processed once in order to avoid repetitive calculations. The second step in the process is the creation of the MSAs. Using PSI-BLAST we find potential homologues in the UniProtKB/SwissProt (15) database using an e-value cutoff of 10−3 and three iterations. The results are forwarded to a filtering script that removes redundant sequences according to three criteria: (i) sequence identity—sequences with more than 95% identity to the query sequence are removed; (ii) sequence length—sequences shorter than 60% of the query sequence are removed; (iii) maximum overlap—since BLAST is a local alignment algorithm, fragment sequences that overlap by over 10% are also removed. Next, redundant sequences are removed using CD-HIT (16); a maximum of the 300 most significant hits (i.e. sequences with the lowest e-values) are selected as homologues, and MUSCLE is used to align them. If a total of less than 50 homologues are found, the entire process is repeated using the Clean_UniProt database. Clean_UniProt is a modified version of the UniProt database (15) aimed to screen the more reliable sequences based on two criteria: (i) if the ‘Description’ (DE) field contain ‘Disease’, ‘RIKEN’, ‘variant’, ‘mutation’, ‘mutant’ or ‘whole genome shotgun sequence’ the sequence is removed; (ii) if the database is ‘TrEMBL’ and the ‘Comments’ (CC) lines contain the word ‘CAUTION’ the sequence is removed. The Clean_UniProt includes non-reviewed entries and is about 10 times larger than UniProtKB/SwissProt. The number of chains supported by each sequence database is presented in Table 2 of the supporting materials.
The third and most CPU-bound step is the execution of Rate4Site to produce the evolutionary scores for each amino acid position in the protein. A Condor (17) job system that is part of the European grid network was used to this end, which allowed us to complete this part of the process for all the polypeptide chains in the PDB within less than 5 days. Rate4Site output includes a Newick formatted phylogenetic tree of the homologues and a list of conservation scores for each of the amino acids positions in the original sequence.
The last step includes parsing of the Rate4Site scores and formatting them to create a range of output data. The scores are normalized and classified into nine conservation levels, as explained in the Introduction section above. These levels are subsequently used for visualization (e.g. Figure 1), using RasMol (18) coloring script and FirstGlance in Jmol. The confidence interval, which is assigned to each amino acid position, represents the reliability of the conservation score of that position. For example, a conservation score for a position that consists mostly of gaps will have a large confidence interval, i.e. low reliability. Low reliability positions are marked yellow in the 3D visualization (2).
All data including intermediate calculations are saved in each chain's directory and a user-friendly HTML page is created to allow viewing the results using a web browser.
ConSurf-DB IN NUMBERS
The build statistics for the first full version of the ConSurf-DB database are presented in Table 1. The initial version of ConSurf-DB was built based on a PDB containing 48 091 entries, using PSI-BLAST v2.2.14 on UniProtKB/SwissProt v54.6 and Clean_UniProt containing 4 225 158 sequences. A total of 117 384 chains were found, 30 918 of which were unique amino acid polypeptides, conforming to our length and modification percentile requirements.
Table 1.
Build statistics for the first full version of ConSurf-DB dated February 2008
| PDB chains | MSA sizes | ||
|---|---|---|---|
| PDB entries processed | 48 091 | Chains with less than 5 homologues (insufficient) | 1348 |
| Total chains found | 117 384 | MSAs Created | 29 570 |
| Filtered | Chains with 5-10 homologues | 859 | |
| Chains containing nucleic acids | 8237 | Chains with 11-20 homologues | 1059 |
| Chains of less than 30 residues | 5594 | Chains with 21-50 homologues | 2332 |
| Chains containing more than 15% modifications | 281 | Chains with 51-100 homologues | 7297 |
| Total chains meeting our requirements | 103 272 | Chains with 101-200 homologues | 14 945 |
| Total distinct chains meeting our requirements | 30 918 | Chains with 201-300 homologues | 3078 |
At peak level, the build process was using 70–150 CPUs, ranging from Pentium III to Xeon. The total CPU time for building the entire ConSurf-DB database was ∼14 000 CPU hours with an average CPU time of ∼30 min per chain.
EXAMPLE AND COMPARISON
The cytochrome c protein (PDB ID: 5cyt) can be found in many species including plants, animals and unicellular organisms. It comprises a single polypeptide chain and is absorbed on the inner membrane of the mitochondrion. It participates in the electron transport chain by carrying one electron using its HEME prosthetic group. There are many cytochrome c homologues in UniProtKB/SwissProt and many of them are highly similar to each other. Therefore, the MSA that was constructed using the default parameters of the ConSurf server includes 50 homologues of over 75% sequence identity to the query with an average of 83% and SD of 3.3%. Thus, using this default means of collecting homologues, the vast majority of the residues appeared to be invariant, and was assigned the highest conservation level (Figure 1B, maroon). Additionally, in some of the residues the data was considered as insufficient (Figure 1B, yellow). Overall, the results were unsatisfactory.
The approach that was used to create the ConSurf-DB managed to deal much better with cytochrome c and its ample homologues. From over 180 similar sequences that were found in UniProtKB/SwissProt, 123 were selected by the ConSurf-DB filtering process as homologues to be aligned and analyzed. Sequence identity to the query ranged from 22% to 91% with an average of 58% and SD of 17.6%. Thus, the evolutionary profile obtained makes much more sense in view of the protein function: Highly conserved residues delineate the HEME binding site and no position in the MSA was classified as insufficient (Figure 1A). The MSA in ConSurf-HSSP for cytochrome c shows similar sequence identity values.
Comparison with results from the ‘Evolutionary Trace Viewer’, ‘SiteFiNDER|3D’ and ‘HotPatch’ servers can be found in Figure 3 of the supporting materials.
CONCLUSIONS
ConSurf-DB is a new addition to the ConSurf set of online tools for creating evolutionary conservation profiles of proteins. In most cases it gives better results than the ordinary ConSurf running with default parameters due to the more advance homologues selection process. Moreover, since all the data is pre-calculated there is no waiting time. This makes ConSurf-DB a preferred tool for initial investigation of proteins. The evolutionary profile of the protein can be correlated with results obtained using other computational tools and experimental data to gain functional insight. The conservation profiles can also be linked to other online web servers.
It is important to note that the quality of the results of any evolutionary algorithm depends on the amount of homologous proteins and their diversity over the phylogenetic tree. For that reason the ConSurf-DB repository will be rebuilt annually to incorporate new homologues, which were sequenced during the year.
The automatic procedure that was used here represents a quasi-optimum with regards to the search for homologous proteins and their alignment. However, very often manual intervention can be used to improve this process further, especially when conducted by an expert on a specific protein. Thus, users may still prefer to use the original ConSurf server that allows inputting custom MSAs and phylogenetic trees, as well as changing key parameters.
Generally speaking, functional regions are highly conserved. However, it is noteworthy that there are exceptions to this rule. One particularly interesting case is the recognition region in antibodies and MHC molecules, which are hyper-variable (19). The ConSurf-DB can be used to recognize these regions as well, if the user knows what to look for.
It is also important to notice that in some cases we had to abort the ConSurf analysis of chains not conforming to our basic thresholds of length, modifications percentile and the number of homologues found. One of the key reasons for that was an insufficient number of homologues. It is anticipate that as the various genome and meta-genome projects are moving forward and sequences accumulate, we will be able to cover the entire PDB. Until then, complementary tools, such as THEMATICS (20) and HotPatch, may be used to find functional regions without the need to look for homologous proteins.
We are hopeful that ConSurf-DB will be a valuable tool for researchers and anticipate that it will assist in the discovery of protein function. To this end, we are constantly working on adding ConSurf-DB results to online protein databases. The PDBsum (21) database will present direct links to ConSurf-DB and the Proteopedia Project (22) will integrate the ConSurf-DB data, allowing users to browse conservation scores without leaving the site.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the German Federal Ministry of Education and Research (BMBF) within the framework of German-Israeli Project Cooperation (DIP) (grant number K5.1). Funding for open access charge: The German Federal Ministry of Education and Research (BMBF) within the framework of German-Israeli Project Cooperation (DIP) (grant number K5.1).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Eric Martz for his constant feedback on the various ConSurf tools. This feedback was reflected in the design of ConSurf-DB.
REFERENCES
- 1.Madabushi S, Yao H, Marsh M, Kristensen DM, Philippi A, Sowa ME, Lichtarge O. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J. Mol. Biol. 2002;316:139–154. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- 2.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 4.Glaser F, Rosenberg Y, Kessel A, Pupko T, Ben-Tal N. The ConSurf-HSSP database: the mapping of evolutionary conservation among homologs onto PDB structures. Proteins. 2005;58:610–617. doi: 10.1002/prot.20305. [DOI] [PubMed] [Google Scholar]
- 5.Dodge C, Schneider R, Sander C. The HSSP database of protein structure-sequence alignments and family profiles. Nucleic Acids Res. 1998;26:313–315. doi: 10.1093/nar/26.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayrose I, Graur D, Ben-Tal N, Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol. Biol. Evol. 2004;21:1781–1791. doi: 10.1093/molbev/msh194. [DOI] [PubMed] [Google Scholar]
- 7.Morgan DH, Kristensen DM, Mittelman D, Lichtarge O. ET viewer: an application for predicting and visualizing functional sites in protein structures. Bioinformatics. 2006;22:2049–2050. doi: 10.1093/bioinformatics/btl285. [DOI] [PubMed] [Google Scholar]
- 8.Innis CA. siteFiNDER|3D: a web-based tool for predicting the location of functional sites in proteins. Nucleic Acids Res. 2007;35:W489–W494. doi: 10.1093/nar/gkm422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettit FK, Bare E, Tsai A, Bowie JU. HotPatch: a statistical approach to finding biologically relevant features on protein surfaces. J. Mol. Biol. 2007;369:863–879. doi: 10.1016/j.jmb.2007.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 17.Thain D, Tannenbaum T, Livny M. Distributed computing in practice: the Condor experience. Concurr. Pract. Exper. 2005;17:323–356. [Google Scholar]
- 18.Bernstein HJ. Recent changes to RasMol, recombining the variants. Trends Biochem. Sci. 2000;25:453–455. doi: 10.1016/s0968-0004(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 19.Reche PA, Reinherz EL. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 2003;331:623–641. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 20.Ko J, Murga LF, Wei Y, Ondrechen MJ. Prediction of active sites for protein structures from computed chemical properties. Bioinformatics. 2005;21(Suppl. 1):i258–i265. doi: 10.1093/bioinformatics/bti1039. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodis E, Prilusky J, Martz E, Silman I, Moult J, Sussman JL. Proteopedia – a scientific ‘wiki’ bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biol. 2008;9:R121. doi: 10.1186/gb-2008-9-8-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]