Abstract
The increasing structural information about target-bound compounds provide a rich basis to study the binding mechanisms of metabolites and drugs. SuperSite is a database, which combines the structural information with various tools for the analysis of molecular recognition. The main data is made up of 8000 metabolites including 1300 drugs, bound to about 290 000 different receptor binding sites. The analysis tools include features, like the highlighting of evolutionary conserved receptor residues, the marking of putative binding pockets and the superpositioning of different binding sites of the same ligand. User-defined compounds can be edited or uploaded and will be superimposed with the most similar co-crystallized ligand. The user can examine all results online with the molecule viewer Jmol. An implemented search algorithm allows the screening of uploaded proteins, in order to detect potential drug binding sites, which are similar to known binding pockets. The huge data set of target-bound compounds in combination with the provided analysis tools allow to inspect the characteristics of molecular recognition, especially for drug target interactions. SuperSite is publicly available at: http://bioinformatics.charite.de/supersite.
INTRODUCTION
The Protein Data Bank (1) contains crystallographic information about proteins, which are co-crystallized with thousands of metabolites or drugs. The data are highly relevant not only for analyzing the recognition of individual compounds (2), but also as a learning set for molecular interaction models (3). In many cases, small compounds bound to macromolecules are medically active and listed as approved drugs. The consideration of such co-crystallized structures can considerably facilitate the process of drug development (4). Another important aspect of molecular interaction is the specificity of a ligand. Many compounds address several receptor proteins. Comparative analysis of the target proteins can enable to draw conclusions about the molecular recognition between ligands and targets (5). One paradigm, that frequently reoccurs, is the concept of structure activity relationship (SAR)—either meaning, that similar ligands have a similar mode of action (6), or that similar binding sites may share binding partners. This paradigm has implications for finding novel leads, as well as the elucidation of possible side effects (7). SitesBase (8) is an excellent source, which utilizes this similarity concept, using an indexing algorithm, that allows fast comparisons of similar binding sites. This enables the researcher to quickly generate hypotheses about probabilities that a certain binding site will be adopted by a ligand. For further investigations of the interactions between small compounds and macromolecules, a variety of additional sources are available. Concerning experimentally available binding data like Kd, Ki and IC50 data, the Binding MOAD (9), PDBbind (10) and the Binding Database (11) are of special interest, since they allow conclusions about the binding affinity of the compounds. Regarding the integration of secondary databases like SCOP (12), CATH (13) and Pfam (14), there is a variety of excellent sources with a strong focus on macromolecules, like PDBsum (15), RCSB PDB (1) and IMB Jena Image Library (16), while PROCOGNATE (17) is especially tailored for elucidating enzymatic activity. However, there is no single resource, which is centered on drug-like compounds, while integrating all available structural information. Therefore, SuperSite was created with three main design goals in mind:
Rich integration of the PDB, including full-text search, complete 3D information and extraction of ligand–receptor relationships.
Integration of secondary sources, to detect putative binding sites.
Detection and highlighting of compounds, which are considered to be medically active.
The aim of SuperSite therefore is, to assist the structural biologist with an online tool, which facilitates the inspection of known and putative binding sites, regarding likely binding sites and conservation information. For drug-like compounds, we are additionally providing superimposed binding sites of the same ligand, which allows the detection of structurally conserved residues.
DATABASE AND TOOLS
Primary database
SuperSite's main data source is the PDB, currently containing over 51 000 3D structures and providing well over 290 000 implicit interactions of macromolecules and small compounds. The raw PDB is parsed and translated automatically into a relational database schema that enables SuperSite to further integrate secondary databases for information enrichment (see subsequent subsection). To make the knowledge in the primary database accessible, SuperSite is providing extensible means for querying. The main text query possibilities include the search for PDB-ID, Het-ID, protein, ligand names and synonyms, as well as a full text search, which screens the complete header of all PDB files for a given term. For instance, searching for the term ‘insulin’ reveals all insulin-related proteins so that they can be used for further investigation. An important subgroup of the proteins in the PDB, are enzymes involved in many catalytic activities. To this end, SuperSite provides an EC tree presentation (18), which makes it possible to browse the PDB via enzyme class/subclass and picking proteins of interest. To investigate the similarity of certain proteins, we have integrated the protein similarity cluster information from the Cd-hit algorithm (18). This information is provided for 95, 90, 70 and 50% similarity, based on the sequence. A specialized search form not only allows the search for similar proteins, but also allows searching for apo-/holo-states. This directly allows to deal with the question, how much the bound form of a protein differs from the unbound form. When it comes to the field of small compounds in the PDB, SuperSite is providing appliances for filtering physio-chemical features like molecular weight, chemical formula or number of atoms. A built-in tool for finding similar small ligands to a given one, is a fingerprint search, based on MyChem fingerprints (http://mychem.sf.net). SuperSite also provides Marvin as an online tool (http://www.chemaxon.com/), which allows to draw or upload a molecule, and screen it against all ligands contained in the PDB (sdf and mol file formats are supported). User-defined compounds are visualized by a superposition according to the most similar bound ligand.
Secondary data sources
To assist the user in investigating potential binding partners and putative binding pockets, SuperSite integrates secondary information from related data sources. Analyses of functionally important sites suggest that the degree of conservation within a protein family is a hint for potential binding sites (19). To this end, SuperSite integrates information from HSSP (20), a data source, which contains information about the degree of residue conservation within a family of proteins. As additional source of information, we are providing de novo predictions of possible protein binding pockets calculated by LIGSITEcsc (21). This information is precalculated and also stored in the database. HSSP and LIGSITEcsc provide exhaustive information about putative binding sites. Together with the possibility to elucidate related proteins (as described in the subsection above), this provides starting points for the detection of putative binding sites.
Drug site encyclopedia
A subset of all relations, between proteins and small compounds, is the relationship of proteins and drugs. This subset is of high importance, when it comes to a systematic investigation of the desired effects of drugs (on- and off-target effects). An implemented part of SuperSite therefore is the Drug Site Encyclopedia. As the term drug is not self-defining, we compared the World Drug Index (http://scientific.thomsonreuters.com), the Comprehensive Medical Chemistry (CMC) Database (http://www.mdl.com), the NCI cancer Compounds (http://dtp.nci.nih.gov) and SuperDrug (22) with all ligands of the PDB to determine the intersection set using standard fingerprints from OpenBabel (http://openbabel.org). The screening was performed via a fingerprint search (http://mychem.sf.net). Entities with a Tanimoto coefficient of >0.85 and an equal number of nonhydrogen atoms were considered as drugs (23). This screening yielded more than 1300 medicinal compounds in the PDB. Within the Drug Site Encyclopedia, we are providing extended instruments for exploring the relationship between drug and target. One aspect is the possibility to investigate the superimposed binding sites of the same ligand, showing residues that are conserved in a spatial region, or frequently occur in a region characteristic for drug recognition. Additionally, we are providing a point set match algorithm, which uses known binding sites (patches) of a ligand, to recognize similar patches on the surface of uploaded structures—solved structures or models (algorithm to be published elsewhere). SuperSite is also calculating Lipinski's Rule Of Five (24), reflecting the drug-likeness of uploaded or edited compounds.
Visualization, browsing and availability
SuperSite can be used with a standard web browser with active Java 1.5+. The molecular viewer Jmol (http://jmol.org) visualizes proteins, ligands and interactively highlights all integrated data sources like HSSP or LIGSITEcsc. SuperSite also allows browsing between ligand and protein interactions, and vice versa. For instance, it is possible to query the protein ‘Insulin’, pick out a ligand and jump to the next view providing all co-crystallized proteins. If the ligand is contained in the Drug Site Encyclopedia, it is also possible to investigate the superimposed binding sites. Links are provided to numerous relevant sources, containing further specialized data sources [e.g. Proteopedia (25), RCSB, PDBSum]. SuperSite is accessible free of charge for academic institutions. Flat files of the database are available upon request.
CASE STUDIES
Case study 1: PLP binding partners and spatial mining
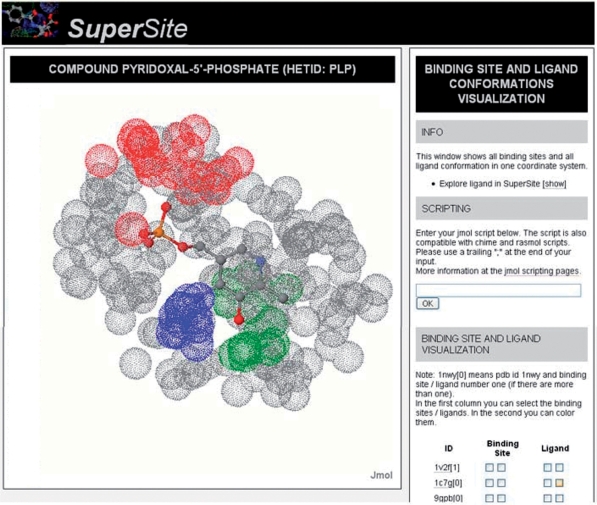
Vitamin B6 (Het-ID: PLP) is a co-enzyme, mainly used in the amino acid metabolism and widely present in the human body. Currently, SuperSite contains information about 463 structures containing PLP, representing, a variety of proteins (e.g. aminotransferases, glycogen phosphorylases). A visualization of all binding sites at once can be achieved, by selecting ‘Drug Encyclopedia’ in the main menu and then entering ‘PLP’ as Het-ID. This view allows inspecting common features, like spatial conservation of specific amino acid types. In the case of PLP, it gets obvious, that, for instance, residue Gly is conserved at a spatial position near the phosphate (Figure 1). This is even the case, when the proteins are structurally dissimilar, a conclusion also discussed in ref. (26).
Figure 1.
Superimposed binding sites of the ligand Vitamin B6 (Het-ID: PLP) from PDB-IDs: 1BJO, 1C7N and 1DJE. Although the proteins show an overall dissimilar structure, residue glycine (red), lysine (blue) and histidine (green) are clustering at specific spatial positions (other atoms of the binding sites depicted in gray).
Case study 2: determination of binding pockets
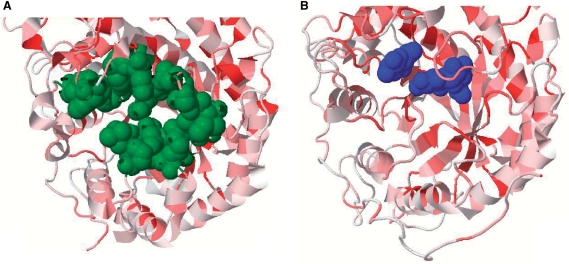
The elucidation of possible binding pockets and active sites of proteins without co-crystallized compounds is a common task for structural biologists. SuperSite provides two tools for the investigation into this topic: LIGSITEcsc—providing precalculated binding pocket predictions and HSSP—providing information about sequence conservation. For instance, PDB-ID 1WDP refers to the structure of the enzyme β-amylase, solved without substrate. To evaluate if there is a possible binding pocket, the user can consult LIGSITEcsc and HSSP interactively from SuperSite (Figure 2). The HSSP conservation shows a more conserved region around residue glutamine (residue number 186). At the same position, LIGSITEcsc shows a relatively large predicted binding pocket. There is another β-amylase (PDB-ID: 1B1Y) similar in overall structure to the apo form containing a ligand at the position proposed by LIGSITEcsc and HSSP what shows the applicability of this method.
Figure 2.
An apo (PDB-ID: 1WDP, A) and holo form (PDB-ID: 1B1Y, B) of β-amylase. The predictions for the binding site pocket (green) as well as the HSSP conservation (red conserved, white not conserved) support the hypothesis of a binding site at this position. This claim can be proved by the holo form (B) with α-d-glucose (blue), bound to the predicted pocket.
Case study 3: detection of binding partners via similarity screening
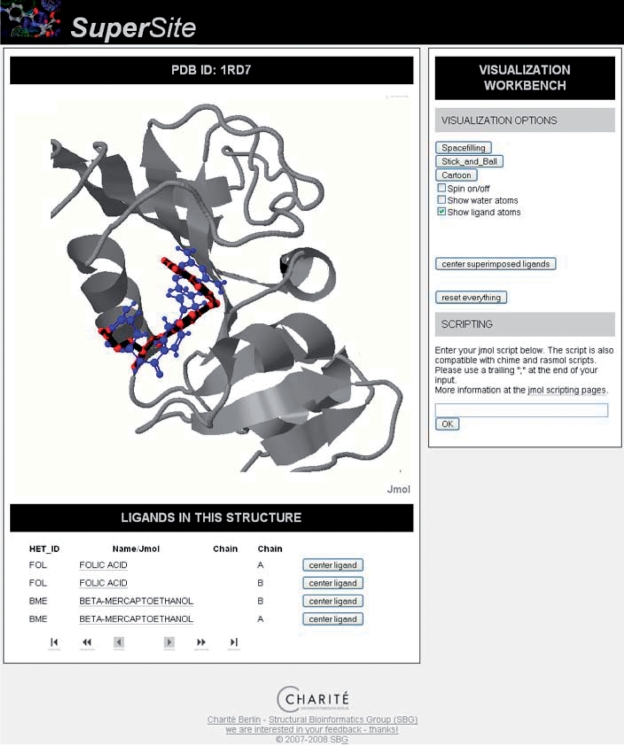
SuperSite also offers a facility for the fast similarity screening of a compound, against all ligands co-crystallized in the PDB. This enables to hypothesize about possible binding partners for similar compounds. Methotrexate (Het-ID: MTX) is a drug, which is used as anti-inflammatory agent/immunosuppressant and in high concentrations used as chemotherapeutical agent (27). Methotrexate inhibits the folic acid biosynthesis and therefore slows the proliferation of cells. SuperSite enables the user to find similar compounds in the PDB, by simply drawing, or by uploading a mol or sdf file. After issuing the similarity search for Methotrexate, one of the best hits not identical to Methotrexate, is folic acid (HET-ID: FOL) bound to a dihydrofolate reductase (Figure 3). The query compound Methotrexate is superimposed with folic acid, which is the known mode of action.
Figure 3.
A dihydrofolate reductase (PDB-ID: 1RA7) with folic acid (HET-ID: FOL, red) bound. One of the highest ranking results from a ligand similarity screening, using compound Methotrexate (Het ID: MTX, blue), suggests a binding at that position.
CONCLUSIONS
We presented a novel database, SuperSite that offers 3D information about proteins and about their bound compounds (ligands). SuperSite enables the user to investigate into the relationship of ligand and receptor in atomic detail, integrating information sources about putative binding sites and conservation on residue level. SuperSite is made with an emphasis on ligands, that are drug-like and therefore of special interest for medical research. To this end, SuperSite provides 3D superpositions of all binding sites of a certain ligand, which enable the user to investigate into the spatial arrangement and properties of the binding site. For further investigations, SuperSite allows to issue a similarity screening against ligands bound in macromolecules as well as a screening of proteins against known binding sites. SuperSite is publicly available at: http://bioinformatics.charite.de/supersite.
FUNDING
Deutsche Krebshilfe, Deutsche Forschungsgemeinschaft (DFG SFB-449); International Research Training Group on Genomics and Systems Biology of Molecular Networks (GRK1360). Funding for open access charge: Deutsche Forschungsgemeinschaft (SFB-449).
Conflict of interest statement. This work is licensed under a Creative Commons Attribution-Noncommercial-Share Alike 3.0 License.
ACKNOWLEDGEMENTS
The authors would like to thank Björn Grüning for maintaining servers and supporting the project quietly, as well as Silvana Gromöller, Maria Krügle and Sabrina Kleeßen for coding parts of the prototype. The authors also thank Andrean Goede and Jessica Ahmed for providing superposition code. Without the use of free and/or open source software this effort would not have been possible. In this regard, the SuperSite Team especially wants to thank Biojava (http://biojava.org), CDK (http://cdk.sf.net), Jmol (http://jmol.org) and MyChem (http://mychem.sf.net).
REFERENCES
- 1.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 3.Schormann N, Senkovich O, Walker K, Wright DL, Anderson AC, Rosowsky A, Ananthan S, Shinkre B, Velu S, Chattopadhyay D Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function. Proteins. 2008. [Epub ahead of print] doi: http://dx.doi.org/10.1002/prot.22115. [DOI] [PubMed]
- 4.Bayry J, Tchilian EZ, Davies MN, Forbes EK, Draper SJ, Kaveri SV, Hill AV, Kazatchkine MD, Beverley PC, Flower DR, et al. In silico identified CCR4 antagonists target regulatory T cells and exert adjuvant activity in vaccination. Proc. Natl Acad. Sci. USA. 2008;105:10221–10226. doi: 10.1073/pnas.0803453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tikhonova IG, Sum CS, Neumann S, Engel S, Raaka BM, Costanzi S, Gershengorn MC. Discovery of novel agonists and antagonists of the free fatty acid receptor 1 (FFAR1) using virtual screening. J. Med. Chem. 2008;51:625–633. doi: 10.1021/jm7012425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunkel M, Gunther S, Ahmed J, Wittig B, Preissner R. SuperPred: drug classification and target prediction. Nucleic Acids Res. 2008;36:W55–W59. doi: 10.1093/nar/gkn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minai R, Matsuo Y, Onuki H, Hirota H. Method for comparing the structures of protein ligand-binding sites and application for predicting protein-drug interactions. Proteins. 2008;72:367–381. doi: 10.1002/prot.21933. [DOI] [PubMed] [Google Scholar]
- 8.Gold ND, Jackson RM. SitesBase: a database for structure-based protein-ligand binding site comparisons. Nucleic Acids Res. 2006;34:D231–D234. doi: 10.1093/nar/gkj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson ML, Smith RD, Khazanov NA, Dimcheff B, Beaver J, Dresslar P, Nerothin J, Carlson HA. Binding MOAD, a high-quality protein-ligand database. Nucleic Acids Res. 2008;36:D674–D678. doi: 10.1093/nar/gkm911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R, Fang X, Lu Y, Yang CY, Wang S. The PDBbind database: methodologies and updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Liu M, Gilson MK. BindingDB: a web-accessible molecular recognition database. Comb. Chem. High Throughput Screen. 2001;4:719–725. doi: 10.2174/1386207013330670. [DOI] [PubMed] [Google Scholar]
- 12.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn R, Griffiths-Jones S, Bateman A. Curr. Protoc. Bioinformatics. 2003. Identifying protein domains with the Pfam database. Chapter 2, Unit 2.5. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichert J, Suhnel J. The IMB Jena Image Library of Biological Macromolecules: 2002 update. Nucleic Acids Res. 2002;30:253–254. doi: 10.1093/nar/30.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashton M, Nobeli I, Thornton JM. PROCOGNATE: a cognate ligand domain mapping for enzymes. Nucleic Acids Res. 2008;36:D618–D622. doi: 10.1093/nar/gkm611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett AJ. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). Enzyme nomenclature. Recommendations 1992. Supplement 3: corrections and additions (1995) Eur. J. Biochem. 1996;237:1–5. doi: 10.1111/j.1432-1033.1996.t01-1-00001.x. [DOI] [PubMed] [Google Scholar]
- 19.Chakrabarti S, Lanczycki CJ. Analysis and prediction of functionally important sites in proteins. Protein Sci. 2007;16:4–13. doi: 10.1110/ps.062506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodge C, Schneider R, Sander C. The HSSP database of protein structure-sequence alignments and family profiles. Nucleic Acids Res. 1998;26:313–315. doi: 10.1093/nar/26.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct. Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goede A, Dunkel M, Mester N, Frommel C, Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 23.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 24.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 25.Hodis E, Prilusky J, Martz E, Silman I, Moult J, Sussman JL. Proteopedia - a scientific ‘wiki’ bridging the rift between 3D structure and function of biomacromolecules. Genome Biol. 2008;9:R121. doi: 10.1186/gb-2008-9-8-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kume A, Koyata H, Sakakibara T, Ishiguro Y, Kure S, Hiraga K. The glycine cleavage system. Molecular cloning of the chicken and human glycine decarboxylase cDNAs and some characteristics involved in the deduced protein structures. J. Biol. Chem. 1991;266:3323–3329. [PubMed] [Google Scholar]
- 27.Green MR, Chamberlain MC. Renal dysfunction during and after high-dose methotrexate. Cancer Chemother. Pharmacol. 2008. [Epub ahead of print] doi: http://dx.doi.org/0.1007/s00280-008-0772-0. [DOI] [PubMed]