Abstract
Influenza epidemics arise through the acquisition of viral genetic changes to overcome immunity from previous infections. An increasing number of complete genomes of influenza viruses have been sequenced in Asia in recent years. Knowledge about the genomes of the seasonal influenza viruses from different countries in Asia is valuable for monitoring and understanding of the emergence, migration and evolution of strains. In order to make full use of the wealth of information from such data, we have developed an integrated user friendly relational database, Influenza Sequence and Epitope Database (ISED), that catalogs the influenza sequence and epitope information obtained in Asia. ISED currently hosts a total of 13 020 influenza A and 2984 influenza B virus sequence data collected in 17 countries including 9 Asian countries, and a total of approximately 545 amantadine-resistant influenza virus sequences collected in Korea. ISED provides users with prebuilt application tools to analyze sequence alignment and different patterns and allows users to visualize epitope-matching structures, which is freely accessible at http://influenza.korea.ac.kr and http://influenza.cdc.go.kr.
INTRODUCTION
Influenza is one of the most important respiratory infectious diseases of humans. It is estimated that influenza is responsible for 250 000–500 000 deaths annually (1). The 1918 pandemic resulted in the deaths of 20–50 million on a global scale, which was one of the most devastating disease outbreaks in human history (2). Influenza viruses of the family Orthomyxoviridae contain eight single-stranded negative-sense RNA molecules which encode a total of 11 proteins. Three antigenically distinct virus types—A, B and C—circulate in human populations (3). Antigenic drift of the viruses makes the existing vaccines ineffective and antigenic shift creates new strains which may cause worldwide pandemic. Genome sequences of currently circulating virus isolates are important sources of information about influenza. Recent developments in viral genome sequencing, antigenic mapping and epidemiological modeling are greatly improving our knowledge of the evolution of human influenza virus (4–6). However, many aspects of the evolutionary and epidemiological dynamics of influenza viruses are still far from complete.
Significant efforts have been made to build public resources of influenza viruses, such as the Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) at the National Center for Biotechnology Information (NCBI), the Influenza Sequence Database at Los Alamos National Laboratory, the Influenza Virus Database (http://influenza.genomics.org.cn) at the Beijing Institute of Genomics and the BioHealthBase Bioinformatics Resource Center (http://www.biohealthbase.org) (7–10). An increasing number of genomes of influenza viruses have been sequenced in Asia in recent years. Southern China has long been considered a potential epicenter for emergence of pandemic influenza viruses (11) and becomes one of the major foci for viral surveillance. Tropical regions may function as permanent mixing pools for viruses from around the world, providing ideal source populations because of extended viral transmission (12). Knowledge about the genomes of the seasonal influenza viruses from different countries in Asia is valuable for monitoring and understanding of the evolution and migration of strains. Since 1968, Korea National Institute of Health (KNIH) has performed influenza virus isolation as part of the World Health Organization's influenza surveillance network. In 2000, the Korean Influenza Surveillance Scheme was established as an integrated clinical and laboratory surveillance network involving public health centers and private clinics (13). Sentinel physicians report cases of influenza-like illness weekly and forward specimens to KNIH for virus isolation and characterization. KNIH has sequenced the isolates of influenza viruses collected in Korea, which have been registered to GenBank at the NCBI.
New insights into immunity initiated by host–pathogen interaction are changing the way we think about pathogenesis of influenza. The immune response to influenza virus infection is directed against various epitopes of antigens. Two important surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) mutate at high frequencies under the strong selective pressure of the host's immune system (14). Epitopes can be used to monitor immune response and a single amino acid mutation at the key residue of the epitope is frequently sufficient to cause an antigenic change (15). High-level antiviral drug resistance can also be conferred by single amino acid substitutions (16). Over the years, influenza antiviral drug resistance has grown rapidly despite the efficacy of the drugs comparable to that of vaccines. In order to leverage the wealth of information from such data, we have developed an integrated user friendly relational database, Influenza Sequence and Epitope Database (ISED), particularly focusing on the genomes of the seasonal influenza viruses from Asian countries. We have added value by implementing a suite of bioinformatics tools that can be used to analyze and visualize the influenza data. This freely accessible resource will augment influenza research and contribute to improved public heath.
OVERVIEW OF THE DATABASE
ISED was designed to collect, store and provide sequence information on influenza viruses including drug-resistant strains, conjoined to research tools for sequence pattern and epitope structural analyses of the data. At present, ISED includes information on 16 004 influenza sequences (13 020 influenza A and 2984 influenza B viruses) including those from nine Asian countries (China, Japan, Korea, Malaysia, Philippines, Singapore, Taiwan, Thailand and Vietnam) (Table 1). It also hosts 545 drug-resistant influenza sequences against amantadine collected in Korea (Table 2). No drug-resistant influenza isolates were found in Korea against oseltamivir and zanamivir. Influenza virus sequences collected in Korea are registered and will be registered to GenBank at the NCBI immediately upon publication (currently an additional 184 segment sequences as well as 545 drug-resistant sequences). Those of other countries are collected by searching from NCBI GenBank database. ISED also contains a total of 179 T cell epitopes and 5 antibody epitopes experimentally determined or curated from scientific literature, useful for epitope matching.
Table 1.
Current number of influenza virus sequence data in ISED
| Nation | Host | Segmenta |
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M1/M2 | NS1/NS2 | |||
| Australia | Human | 103/4 | 93/4 | 211/4 | 259/70 | 106/4 | 219/9 | 0 | 0 | 991/95 |
| Canada | Human | 4/3 | 4/3 | 4/3 | 7/4 | 2/3 | 49/5 | 0 | 0 | 70/21 |
| China | Human | 32/22 | 33/22 | 32/23 | 810/164 | 45/22 | 343/49 | 78/42 | 67/40 | 1440/384 |
| France | Human | 1/1 | 1/1 | 1/1 | 205/6 | 1/1 | 33 | 0 | 0 | 242/10 |
| Germany | Human | 33/0 | 17/0 | 28/0 | 116/1 | 33/0 | 35/0 | 0 | 0 | 262/1 |
| Italy | Human | 0/2 | 0/1 | 0/2 | 91/86 | 0/2 | 4/40 | 0 | 0 | 95/133 |
| Japan | Human | 14/40 | 14/40 | 14/40 | 655/200 | 15/48 | 15/52 | 32/89 | 29/109 | 788/615 |
| Korea | Human | 3/0 | 3/0 | 3/0 | 265/81 | 3/0 | 41/5 | 48/2 | 6/2 | 372/90 |
| Malaysia | Human | 0 | 0 | 0 | 59/22 | 0 | 0 | 0 | 0 | 59/22 |
| Philippines | Human | 0 | 0 | 0 | 66/60 | 0 | 0 | 0 | 0 | 66/60 |
| Singapore | Human | 0 | 0 | 0 | 86/13 | 0 | 0 | 0 | 0 | 86/13 |
| Spain | Human | 0/1 | 0/1 | 0/1 | 72/23 | 0/1 | 6/3 | 0 | 0 | 78/30 |
| Taiwan | Human | 6/2 | 6/2 | 6/2 | 254/336 | 6/2 | 6/2 | 12/12 | 11/11 | 307/354 |
| Thailand | Human | 0/6 | 0/6 | 0/6 | 124/70 | 0/6 | 0/12 | 0/12 | 0/11 | 124/129 |
| USA | Human | 1277/120 | 1062/120 | 1386/117 | 1799/272 | 1347/120 | 908/264 | 0 | 0 | 7779/1013 |
| United Kingdom | Human | 12/1 | 11/1 | 8/1 | 123/9 | 15/1 | 15/1 | 0 | 0 | 184/14 |
| Vietnam | Human | 0 | 0 | 0 | 77/0 | 0 | 0 | 0 | 0 | 77/0 |
| Total | 1485/202 | 1244/201 | 1693/200 | 5068/1417 | 1573/210 | 1674/442 | 170/149 | 113/163 | 13 020/2984 | |
aInfluenza virus type A/B.
Table 2.
Current number of drug-resistant virus data in Korea
| Amantadine-resistant influenza virus strains in Korea | ||||||
|---|---|---|---|---|---|---|
| Season | A/H1N1 |
A/H3N2 |
||||
| Total number of isolates | Resistant/ tested | Percent resistance | Total number of isolates | Resistant/ tested | Percent resistance | |
| 2003–2008a | 1858 | 156/302 | 51.7 | 4418 | 389/684 | 56.9 |
| Oseltamivir/zanamivir-resistant influenza virus strains in Korea | |||||||
|---|---|---|---|---|---|---|---|
| Season | A/H1N1 |
A/H3N2 |
B |
Percent resistance | |||
| Total number of isolates | Resistant/ tested | Total number of isolates | Resistant/ tested | Total number of isolates | Resistant/ tested | ||
| 2002–2007b | 1276 | 0/105(60) | 4938 | 0/683(244) | 1434 | 0/146(32) | 0 |
aThe 2008 data included the number of isolates determined by the 7th week.
bThe values in parentheses represent the number of isolates against zanamivir.
The data are updated on a regular basis by a curation team, composed of researchers at the Center for Infectious Diseases at KNIH and in Korea University, in order to ensure a consistently high data quality. The data in ISED is open and freely accessible to the general public, which is one of the chief goals of ISED to offer users easy Web access and graphical user interfaces. ISED is a part of the National BioBank project intended to integrate a framework for identifying, collecting, distributing and managing of biomateirals, which is being developed at KNIH.
DATABASE DESIGN AND CONTENTS
The virus sequences in ISED are categorized into tables according to countries, each of which is characterized by a number of attributes: strain name, target host, virus type, virus subtype or lineage (B type only), RNA segment, amino acid sequence, start number of amino acid sequence, aligned amino acid sequences, NCBI accession number (amino acid sequence), nucleotide sequence, start number of nucleotide sequence, aligned nucleotide sequences, NCBI accession number (nucleotide sequence), reference, author list, isolated region, isolated year and isolated season, followed by oseltamivir/zanamivir-resistant and amantadine-resistant viral sequences if available (data not shown). Reference, one of the attributes, is linked to the PubMed abstract and in some instances to the full text of the article if the journal is available online. Target host and isolated region (nation) tables have one-to-many relationships with the virus sequence table, which are frequently used to extract statistical information. Both vaccine and drug-resistant strain sequences are included in the sequence table. The sequences of 46 vaccine strains (9 strains in A/H1N1, 23 in A/H3N2 and 14 in B) are separately grouped as a vaccine strain table. Since 2002, drug-susceptibility surveillance has been routinely undertaken in the characterization of influenza virus isolates submitted to KNIH. Earlier surveillance showed a low incidence of resistance to amantadine (below 10%). However, as of August 2008, 156 amantadine-resistant influenza sequences in A/H1N1 and 389 amantadine-resistant strain sequences in A/H3N2 were collected in Korea (Table 2).
Epitope data were obtained from the Immune Epitope Database and Analysis Resource (IEDB) (http://www.immuneepitope.org/home.do) with 14 reference strain data (15). The database fields in the epitope data table contain epitope residue, start residue number of epitope, number of residues (only B cell response), virus strain, source protein, protein sequence, start residue number of source protein, epitope type (T cell, B cell response or MHC binding), NCBI accession number of source protein and reference. A reference strain table has one-to-many relationship with the epitope table (data not shown).
DATA RETRIEVAL AND TOOLKIT
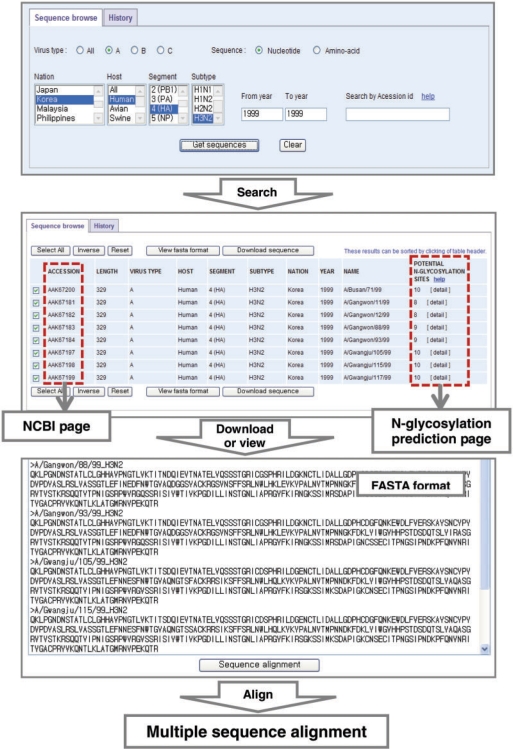
ISED consists of a framework for advanced web-based retrieval, analysis and visualization of related influenza data: sequence browse, sequence analysis and epitope matching arranged in one Oracle schema (17). Sequence data can be retrieved efficiently through establishment of the sequence browse mode (Figure 1). Users can combine various options, such as virus type, nation, host, RNA segment, subtype and collection year. The website then provides access to individual influenza sequence records characterized by a number of database fields, such as accession number, sequence length, virus type, target host, RNA segment, subtype, collected nation and year, virus name and potential N-glycosylation site. The sequence browse results are displayed in chronological order and can also be sorted by column by clicking the table header. Two different search options are provided: individual and collective selection. The amino acid sequences of the selected strains in the displayed list can be retrieved in a separate window by clicking the ‘View fasta format’ button, or can be easily downloaded (Figure 1). Users can prepare an input data by clicking ‘Sequence alignment’ and conduct multiple sequence alignment by direct submission or upload a file of the chosen sequences to CLUSTALW tool of EBI (18). Later, a user's past search history can be located and accessed by the Web server. Drug-resistant influenza sequences can also be retrieved in a sequence browse mode, where alignment or difference patterns of selected resistant sequences can be examined (Figure 2A).
Figure. 1.
Snapshots showing the interrelation of data retrieval tools in ISED. Users can access the data through search options and the results can be selectively saved. The results can be subjected to further analysis, such as multiple sequence alignment.
Figure 2.
The mutant and epitope analysis tools (A) Mutant sequence search, alignment and difference patterns are shown. (B) Epitope sequence and structure analysis showing sequence alignment with reference strains, sequence-matching frequency and an epitope 3D conformation superimposed on a HA structure viewed with Jmol.
The contents of the epitope resource can be searched via user-friendly interface. For epitope matching, users can select virus subtype and reference strains in the reference table containing database fields, such as virus strain, collected area, virus type and target host. Search can query via one of strains and locations which can be selected from pull-down menus, or users can upload and submit their own sequences (Figure 2B). Details of epitope information can be viewed with the strings of amino acid sequences highlighted either in green or blue for antibody or T cell epitopes, respectively. More detailed information can be retrieved by clicking each epitope segment. The epitope 3D structure is visualized using an interactive Jmol (http://www.jmol.org), which is superimposed on an HA tertiary structure model provided by the Protein Data Bank (19). Users can also easily examine matching frequencies between the selected strain and reference strains.
DATA ANALYSIS
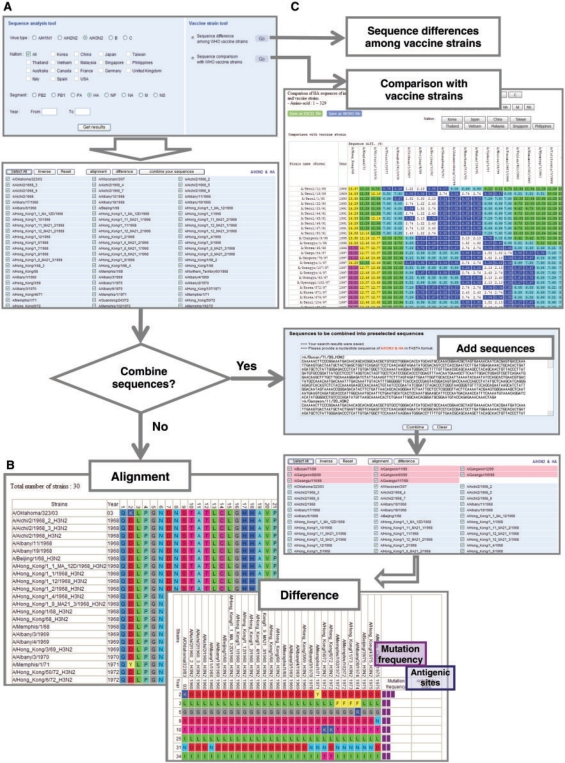
ISED allows access to sequence analysis tools by clicking ‘Sequence analysis’ on the top menu bar. Users can select virus sequence resources via a graphical interface according to virus type, collected region (nation) and RNA segment, with collection year range (Figure 3A), and conduct sequence alignment or sequence difference by clicking the ‘alignment’ or ‘difference’ button. Users can also combine sequences from different sources. On the result page, the alignment can be viewed with color-coded amino acids, so that viral mutations can be seen as changes in color when scanning from the N- to C-terminus along the sequence (Figure 3B). Difference can be also viewed in a separate full-screen window with color coding, where the amino acids with mutations are displayed with additional information of mutation frequencies as well as antigenic sites. Notably, the sequence analysis also includes a vaccine strain tool, with which users can conduct either sequence difference or sequence comparison with vaccine strains (Figure 3C). Sequence difference among vaccine strains returns a result of list of the differences in amino acid sequences among the vaccine strains with information of mutation frequencies as well as antigenic sites. More interestingly, the results of sequence comparison between a sample of circulating strain and vaccine strains can be illustrated by the changes in color patterns against vaccine strains (the lighter the color, the lower the difference). Thus, ISED provides a convenient tool for evaluating the relative closeness of the currently circulating strain against known vaccine strains. Users can then export the results as an Excel or Word file.
Figure 3.
Snapshots showing the interrelation of ISED sequence analysis tools. (A) Users can select sequences or combine sequences from different sources and (B) conduct sequence alignment or sequence comparisons. (C) Sequence differences with vaccine strains are displayed in a separate window with color coding.
FUTURE DIRECTIONS
It is still unclear what features of the influenza viruses are responsible for the global spread and more specifically how the dominant strain is derived. For instance, A/Fujian/411/02 collected in southern China is believed to cause significant outbreaks in China, Japan and Korea in 2002 and spread worldwide during the successive winter season of 2003–2004. The main challenge in the future is to keep ISED up to date with the growing number of complete influenza virus sequences experimentally verified and registered to other databases such as NCBI GenBank. We will thus implement text mining support for database curation in the near future. Toward this goal, a network of influenza expert groups at the Center for Infectious Diseases at KNIH and at Korea University and advisory committee outside KNIH will coordinate validation of new virus strains.
Another challenge is to provide ISED with regional epidemiological features of drug-resistant viruses. Amantadine and rimantadine have been used for the prevention and treatment of influenza A virus infection for >30 years (20). Widespread use of antiviral drugs relying on pandemic stockpiles has the potential to promote emergence of resistant strains of which the epidemiological surveillance is a key to monitor and control. Open sharing of the resistant viral genome information has become increasingly important in preventing and controlling the spread of the resistant viruses. In addition, an antiviral drug resistance analysis tool can be developed and linked to the records in the database, which provides users to analyze influenza sequences for mutations known to confer drug resistance or sensitivity.
The recent H5N1 outbreaks in Asia and a worst outbreak in Korea in 2008 have spurred our interest in surveillance among wild and domestic birds. Avian influenza surveillance may provide early warning signals for any possible introduction of avian viruses in new regions. Importantly, a large number of genome sequences of avian influenza viruses are accumulated in Asia. Given the regions potential as an epicenter for emergence of new influenza virus strains, we particularly intend to extend the ISED platform to enable epidemiological monitoring of avian influenza virus sequences.
USER MANAGEMENT
The ISED management system allows users to access the influenza virus sequence database without registration, except for drug-resistant virus sequence data. However, user registration is required for adding and editing database contents, and user support can be obtained by e-mailing graduate@korea.ac.kr or khkim@korea.ac.kr. Readers are encouraged to contact us if they wish to provide new data for inclusion in ISED, assist with curation or have any suggestions for improvements.
IMPLEMENTATION
ISED was developed as a relational database using Oracle 10g applications (14) on the Windows operating system. Two open source programs, the Apache HTTP Server and Apache Tomcat, were used as HTTP server and servlet container for web service, respectively. Perl scripts were used to provide common gateway interface for sequence alignment using ClustalW, and Java applet was used to link Jmol for displaying 3D models. ISED can be publicly accessed from any Web browser at http://influenza.korea.ac.kr.
FUNDING
Korea National Institute of Health (2008-E00179); BioGreen 21 program grant (20080401-034-008) and the Basic Research Program of the Korea Science & Engineering Foundation. Funding for open access charge: Korea National Institute of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to acknowledge the technical support from Mr C. H. Gong at the Department of Biotechnology & Bioinformatics and Mr J. H. Yeom of the E-Front, Seoul, Korea.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Stohr K. Influenza – WHO cares. Lancet Infect. Dis. 2002;2:517. doi: 10.1016/s1473-3099(02)00366-3. [DOI] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Reid AH, Janczewski TA, Fanning TG. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza cirus. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1857–1859. doi: 10.1098/rstb.2001.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Ann. Rev. Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 4.Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 5.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 6.Russel CA, Jones TC, Barr IG, Cox NJ, Garten RJ, Gregory V, Gust ID, Hampson AW, Hay AJ, Hurt AC, et al. The global circulation of seasonal influenza A (H3N2) virusese. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 7.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The influenza virus resource at the national center for biotechnology information. J. Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macken C, Lu H, Goodman J, Boykin L. The value of a database in surveillance and vaccine selection. In: Osterhaus ADME, Cox N, Hampson AW, editors. Options for the Control of Influenza IV. Amsterdam: Elsevier Science; 2001. pp. 103–106. [Google Scholar]
- 9.Chang S, Zhang J, Liao X, Zhu X, Wang D, Zhu J, Feng T, Zhu B, Gao GF, Wang J, et al. Influenza virus database (IVDB): an integrated information resource and analysis platform for influenza virus research. Nucl. Acids Res. 2007;35:D376–D380. doi: 10.1093/nar/gkl779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Squires B, Macken C, Garcia-Sastre A, Godbole S, Noronha J, Hunt V, Chang R, Larsen CN, Klem E, Biersack K, et al. BioHealthBase: informatics support in the elucidation of influenza virus host-pathogen interactions and virulence. Nucleic Acids Res. 2008;36:D497–D503. doi: 10.1093/nar/gkm905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong CM, Yang L, Chan KP, Leung GM, Chan KH, Guan Y, Lam TH, Hedley AJ, Peiris M. Influenzaassociated weekly hospitalization in a subtropical city. PLoS Med. 2006;3:485–491. doi: 10.1371/journal.pmed.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JS, Shin KC, Na BK, Lee JY, Kang C, Kim JH, Park O, Jeong EK, Lee JK, Kwon JW, et al. Influenza surveillance in Korea: establishment and first results of an epidemiological and virological surveillance scheme. Epidemio. Infect. 2007;135:1117–1123. doi: 10.1017/S0950268807007820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly JM, Wood JM, Robertson JS. Co-circulation and divergence of human influenza viruses. In: Nicholson KG, Webster RG, Hay AJ, editors. Textbook of Influenza. Oxford: Blackwell Science; 1988. pp. 168–177. [Google Scholar]
- 15.Bui H, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc. Natl Acad. Sci. USA. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, et al. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 17.Stephens SM, Chen JY, Davidson MG, Thomas S, Trute BM. Oracle database 10g: a platform for BLAST search and regular expression pattern matching in life sciences. Nucleic Acids Res. 2005;33:D675–D679. doi: 10.1093/nar/gki114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne P. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, Povinelli L, Cox NJ, Klimov AI. Incidence of adamantine resistance among influenza A (H3N2) virusese isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]