Abstract
VectorBase (http://www.vectorbase.org) is an NIAID-funded Bioinformatic Resource Center focused on invertebrate vectors of human pathogens. VectorBase annotates and curates vector genomes providing a web accessible integrated resource for the research community. Currently, VectorBase contains genome information for three mosquito species: Aedes aegypti, Anopheles gambiae and Culex quinquefasciatus, a body louse Pediculus humanus and a tick species Ixodes scapularis. Since our last report VectorBase has initiated a community annotation system, a microarray and gene expression repository and controlled vocabularies for anatomy and insecticide resistance. We have continued to develop both the software infrastructure and tools for interrogating the stored data.
INTRODUCTION
VectorBase is a genome information system which provides a genome browser for visualizing genome annotations, including DNA and protein alignments, variations, protein feature data and functional data sets, such as microarray expression analysis. We are active in producing genome annotation ourselves but also collaborate with a range of partners including our sister Bioinformatic Resource Center's (1) to incorporate and improve the annotations.
The reduction in cost of sequencing has seen genomes become available for an increasing number of vector species. VectorBase is directly responsible for three mosquito species (Aedes aegypti, Anopheles gambiae and Culex quinquefasciatus) and the tick Ixodes scapularis. We work closely with the genome sequencing centres on the initial annotation and publication of these genomes and then assume responsibility for ongoing re-annotation tasks. A number of other genomes are within scope for VectorBase including the body louse (Pediculus humanus), triatomine bug (Rhodnius prolixus), tsetse fly (Glossina morsitans morsitans) and sand flies (Lutzomyia longipalpis and Phlebotomus papatasi). A full list of VectorBase species and data sets can be accessed on the website (http://www.vectorbase.org/Help/Current_release).
This report highlights the new genomes integrated into VectorBase and some of the new features and improvements that we have added since our last report (2). Users interested in the VectorBase project should visit the main web page or help pages (http://www.vectorbase.org/Help/Main_Page) for more information about the project.
ACCESSING VECTORBASE
VectorBase as a web resource is linked with a number of other databases, most notably the public nucleotide and protein databases. Direct cross-references to the genes, transcripts and proteins exist in the GenBank/EMBL/DDBJ genome assembly records as well as the UniProt protein records, where both An. gambiae and Ae. aegypti are deemed to be complete proteomes. Other resources which use VectorBase data range from large general resources, such as Ensembl (http://www.ensembl.org) and Refseq (http://www.ncbi.nlm.nih.gov/RefSeq) to the more biologically focused proteinase database Merops (http://merops.sanger.ac.uk) and miRNA target predictions in mirBase (http://microrna.sanger.ac.uk). The VectorBase site and wiki resource are indexed by the major search engines allowing users to readily find content of interest.
EXPANDED ROLE OF VECTORBASE
VectorBase is active in all stages of genome analysis including initial annotation of new genome sequences in collaboration with the sequencing centres, such as JCVI and The Broad Institute and subsequent re-annotation using both computational and manual approaches in liaison with the community. Automated annotation using the Ensembl system (3) was undertaken for the new genomes (C. quinquefasciatus, P. humanus and I. scapularis). The process of resolving differences between VectorBase and the partner sequencing centre annotations has been a fruitful task leading to high-quality automated annotation but problems will remain which can only be addressed using further resources (expressed sequence tags or new genome sequences) or through manual appraisal of the automated gene predictions. VectorBase has invested some resource toward the latter and implemented strategies for involving the community in the annotation effort. We have also implemented data mining tools, such as the HMMER package (http://hmmer.janelia.org/) to build profile hidden Markov models from multiple sequence alignments which can then be used for sensitive database searching using statistical descriptions of a sequence families consensus.
EXTENSIVE MANUAL APPRAISAL OF MOSQUITO GENE MODELS
The annotation of the An. gambiae genome is being manually appraised using the GMOD annotation tool Apollo (4). Currently, over 50% of the genome has been completed including the entirety of the chromosome arms 2L, 2R and X. Many loci have been updated to correct systematic errors in the computational annotation; especially in reference to tandem arrays of multi-gene families, gene merges from multiple partial predictions and the removal of suspect predictions likely to be based on transposable element sequences. Manual annotations are stored in a separate CHADO database (5), displayed as a track in the genome browser via DAS (6) and integrated into the main gene build during the next round of re-annotation.
Small-scale manual appraisal of gene predictions has been undertaken for An. aegypti and C. quinquefasciatus as part of the quality control for the gene builds. In the case of C. quinquefasciatus, this revealed at least 1500 predictions which were removed from the CpipJ1.2 dataset. Amongst the deprecated gene predictions were a large set of single exon predictions which had no supporting transcript evidence and no similarity to other mosquito proteomes or any other sequences in the public databases. Expert opinion was that these were erroneous over-prediction by the computation algorithms rather than a large Culex-specific gene family. Efforts such as these highlight our determination to improve gene prediction accuracy through the integration of new data sets and the re-appraisal of the existing prediction set.
COMMUNITY ANNOTATIONS
VectorBase employs community representatives focused around the NIAID-funded species (the three mosquito genomes and I. scapularis). The representatives were hired from within the relevant community and have both biological knowledge of the species and informatics skills. Their role is to liaise with the community providing helpdesk and training capacity, acting as mediators and quality assurance for data submission of gene predictions and as advocates for the user community in the development of the VectorBase resource.
We have developed a Community Annotation Pipeline (CAP) to facilitate community involvement in the curation of these genomes. This system consists of a CHADO database which stores annotations, both from the manual effort within VectorBase and those submitted directly from the community, and a web interface submission tool to upload data. Submitters use a spreadsheet format and can include gene predictions, gene symbols and gene descriptions, and attach GO terms or citations to a gene model. One aspect of the submission system is its ability to align a cDNA sequence to the genome using exonerate (7). The simplicity of the submission process in conjunction with community representative involvement in data quality consistency checks (e.g. does the submitted sequence translate correctly) ensures that any required discussion and error correction happens in a timely manner. Currently, the CAP system contains over 13 000 gene predictions. This system replaces the old Anopheles Symbol Database hosted by Ensembl and interested users can find more information about the CAP on the website (http://www.vectorbase.org/Help/Community_Annotation:Submission_User_Guide).
COMPARATIVE ANALYSIS OF THE MOSQUITO GENOMES
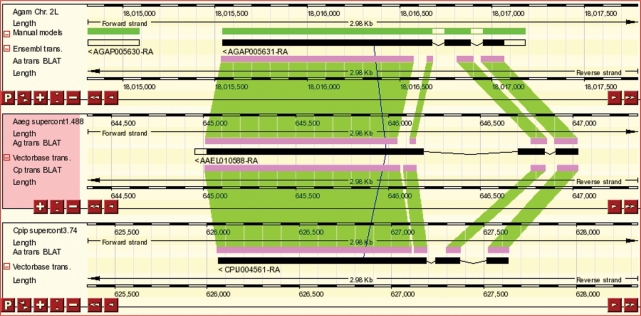
The availability of the ‘Culex’ genome annotation facilitates comparison of the three main families of mosquitoes (Anopheline, Aedine and Culicine) with the model dipteran Drosophila melanogaster. As before VectorBase has calculated pairwise tBLAT alignments (8) that can be used to connect between the genomes in a multi-contig view (Figure 1). Multi-contig views are available using the ‘View alongside’ option in the left-hand navigation panel and as links in the Gene Ortholog section of gene pages. We have added multiple genomic alignments calculated using Pecan (http://www.ebi.ac.uk/~bjp/pecan), which has been shown to be one of the best algorithms in terms of specificity and sensitivity (9). For each genomic position, the level of evolutionary constraint has been evaluated using GERP (10) and stretches of Pecan alignment showing a high level of conservation are marked as constrained elements.
Figure 1.
Three-way comparative display of the mosquito genomes An. gambiae, Ae. aegypti and C. quinquefasciatus (top panel to bottom panel, respectively). Gene predictions are coloured black with the regions of similarity between the genomes indicated by the pink boxes where similarity is indicated by the connection of blocks using the green lines. The upper panel, An. gambiae, also shows a manual annotation confirming the prediction and that the refinement of predictions in one species can be leveraged to improve the quality of prediction across all the mosquito genomes.
Orthologs and paralogs are calculated using Ensembl Compara GeneTrees pipeline. This method is based on maximum likelihood phylogenetic trees built by TreeBest (http://treesoft.sourceforge.net). The trees, presenting the evolutionary history between the genes, are reconciled with the species trees and help in differentiating between duplication and speciation events. The gene tree for a particular gene accessible via the left-hand navigation menu of the gene page and ortholog/paralog data are available for querying via the BioMart interface (11).
GENE EXPRESSION DATABASE
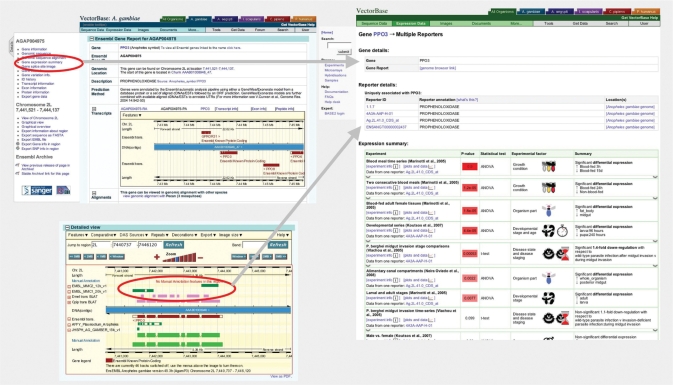
VectorBase has continued to develop its microarray experiment repository and gene expression reports (http://funcgen.vectorbase.org/ExpressionData/). The database now contains 12 array designs (including Affymetrix) and 17 experiments. We continue to actively solicit the community for microarray data which is reflected by the fact that half of the available experiments were published last year. The repository is built around the BASE database system (http://base.thep.lu.se/) and is integrated with the genome via probe mappings in the browser and navigation links for each gene in the GeneView pages (Figure 2). Other new features include text-based search, publication-quality plots and a more robust and extendible implementation of the statistical analysis.
Figure 2.
Integration of the genome browser with the microarray gene expression database. Links from gene pages, highlighted in red in the top left navigation panel, connect to a summary of the gene expression in all available experiments (right-hand panel). Similarly, there are direct links from the mapped reporter probes on the genome browser (bottom left panel) to the gene expression summary based on inferred overlap of the reporter and the current genome annotation. The gene expression summary lists experiments for which the gene has data ranked by statistical significance with a brief summary of the experimental details and links to more detailed analysis.
ONTOLOGIES AND CONTROLLED VOCABULARIES
VectorBase has developed controlled vocabularies describing the anatomy of mosquitoes (Taxonomist's Glossary of Mosquito Anatomy, TGMA), and ticks (Tick Anatomy by Dan Sonenshine, TADS) (12). These ontologies are fully compliant with CARO, the common reference ontology for anatomy (13) and contain 1861 and 628 terms, respectively. The ontologies can be browsed through the website (http://www.vectorbase.org/Search/CVSearch/) allowing for the concurrent visualization of the anatomical structures described. The ontologies are also available from the Open Biomedical Ontologies (OBO) Foundry (http://www.obofoundry.org) which acts as a central repository for all science-based ontologies. The two ontologies enhance the possibilities for annotation of gene expression experiments performed on disease vectors (14).
MIRO and IRBase
AnoBase, the precursor database of VectorBase, has been making available data on insecticide resistance for a number of years (15). VectorBase has expanded this resource, building an ontology (MIRO) that describes features associated with insecticide resistance. This ontology was then used to upgrade the relevant database section with an enhanced search capability. Named IRBase, this can now be searched at http://anobase.vectorbase.org/ir/. Additional data based on new studies as well as on existing published ones are currently being integrated into IRBase, and this tool will be developed into the global database on insecticide resistance for disease vectors.
OUTREACH
VectorBase staff regularly attends teaching workshops to demonstrate the database and the tools available for browsing and query the data. Recent workshops locations include Brazil, Kenya, Mali and South Africa. As the VectorBase genome browser is powered by the Ensembl system their extensive outreach program is also applicable to our databases, potentially giving access to numerous training courses worldwide throughout the year. VectorBase is continuously developing outreach and documentation resources which include the help contact e-mail (info@vectorbase.org) as well as a quarterly newsletter, a FAQ (frequently asked questions), Help Wiki (http://www.vectorbase.org/Help/Main_Page) and a community forum (http://www.vectorbase.org/sections/Forum/index.php). More details relating to these and other resources can be found on the website.
FUTURE DEVELOPMENTS
Massively parallel sequencing technologies (both pyrosequencing and sequencing by synthesis) are being adopted by the sequencing centres reducing the cost of genome sequencing. We expect that this will speed up the generation of genome sequences from vector species which hitherto have been low priority because of size, cost or the practicalities of DNA availability. A number of Anopheles species will be targeted for genome sequencing (http://www.vectorbase.org/Docs/ShowDoc/?doc=WhitePapers) and the reduction in cost means that individual labs can produce significant amounts of sequence data from species or isolates. The integration and management of these data will be a major challenge for the coming years.
Analysis of populations and variation will increase at the sequence level and VectorBase will continue its partnership with Ensembl to process, store and represent this data. Population studies involve other types of data (including insecticide resistance, epidemiology, environmental conditions and vectorial transmission), which are not currently part of the VectorBase data schema and so we will work with partners in the relevant fields to integrate these data with VectorBase enhancing the utility of the resource to the vector genomics community.
FUNDING
NIAID (contract HHSN266200400039C to the core VectorBase project); BioMalPar network of excellence (to the core VectorBase project). Funding for the open access charge: NIAID.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge the many researchers that have provided data through the Community Annotation Pipeline and thank the reviewers for helpful discussions.
REFERENCES
- 1.Greene JM, Collins F, Lefkowitz EJ, Roos D, Scheuermann RH, Sobral B, Stevens R, White O, Di Francesco V. National Institute of Allergy and Infectious Diseases bioinformatics resource centers: new assets for pathogen informatics. Infec. Immun. 2007;75:3212–3219. doi: 10.1128/IAI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2006;35:D503–D505. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D514. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis SE, Searle SMJ, Harris N, Gibson M, Iyer V, Ricter J, Wiel C, Baytraktaroglu L, Birney E, Crosby MA, et al. Apollo: a sequence annotation editor. Genome Biol. 2002;3:R82. doi: 10.1186/gb-2002-3-12-research0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P, Emmert D, Zhang P. Using Chado to store genome annotation data. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0906s12. Chap. 9, Unit 9. [DOI] [PubMed] [Google Scholar]
- 6.Jenkinson AM, Albrecht M, Birney E, Blankenburg H, Down T, Finn RD, Hermjakob H, Hubbard TJ, Jimenez RC, Jones P, et al. Integrating biological data - the distributed annotation system. BMC Bioinformatics. 2008;22:S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent WJ. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper GM, Stone EA, Asimenos G NISC Comparative Sequencing Program, Green ED, Batzoglou S, and Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topalis P, Tzavlaki C, Vestaki K, Dialynas E, Sonenshine DE, Butler R, Bruggner RV, Stinson EO, Collins FH, Louis C. Anatomical ontologies of mosquitoes and ticks, and their web browsers in VectorBase. Insect Mol. Biol. 2008;17:87–89. doi: 10.1111/j.1365-2583.2008.00781.x. [DOI] [PubMed] [Google Scholar]
- 13.Haendel MA, Neuhaus F, Osumi-Sutherland DS, Mabee PM, Mejino JLV, Mungall CJ, Smith B. CARO - the Common Anatomy Reference Ontology. In: Burger A, Davidson D, Baldock R, editors. Anatomy Ontologies for Bioinformatics: Principles and Practice. Chapter 16. New York: Springer; 2008. pp. 311–333. [Google Scholar]
- 14.Topalis P, Lawson D, Collins FH, Louis C. How can ontologies help vector biology? Trends Parasitol. 2008;24:249–252. doi: 10.1016/j.pt.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Topalis P, Koutsos AC, Dialynas M, Kiamos C, Hemingway J, Louis C. AnoBase: a genetic and biological database of anophelines. Insect Mol. Biol. 2005;14:591–597. doi: 10.1111/j.1365-2583.2005.00596.x. [DOI] [PubMed] [Google Scholar]