Abstract
Single nucleotide polymorphisms (SNPs) may be considered the ultimate genetic marker as they represent the finest resolution of a DNA sequence (a single nucleotide), are generally abundant in populations and have a low mutation rate. Analysis of assembled EST sequence data provides a cost-effective means to identify large numbers of SNPs associated with functional genes. We have developed an integrated SNP discovery pipeline, which identifies SNPs from assembled EST sequences. The results are maintained in a custom relational database along with EST source and annotation information. The current database hosts data for the important crops rice, barley and Brassica. Users may rapidly identify polymorphic sequences of interest through BLAST sequence comparison, keyword searches of annotations derived from UniRef90 and GenBank comparisons, GO annotations or in genes corresponding to syntenic regions of reference genomes. In addition, SNPs between specific varieties may be identified for targeted mapping and association studies. SNPs are viewed using a user-friendly graphical interface. The database is freely accessible at http://autosnpdb.qfab.org.au/.
INTRODUCTION
Molecular genetic markers describe genetic variations and provide a link between observed phenotypes and the underlying genotype. The development of high-throughput methods for the detection of single nucleotide polymorphisms (SNPs) and small insertion/deletions (indels) has led to a revolution in their use as molecular markers. SNPs may be considered the ultimate genetic marker as they represent the finest resolution of a DNA sequence, are generally abundant in populations and have a low mutation rate (1). However, SNP markers can be costly to develop, especially where re-sequencing from multiple individuals is required. The mining of readily available sequence data significantly reduces the costs associated with SNP discovery (2). The principal challenge in SNP discovery remains the discrimination between true genetic polymorphisms and the often more abundant sequence errors. Where sequence trace files are available to filter polymorphisms in traces of dubious quality, these can be used to differentiate between true SNPs and sequence error (3). Where trace files are unavailable, the identification of sequence errors can be based on two further methods to determine SNP confidence: redundancy of the polymorphism in a sequence alignment, and co-segregation of putative SNPs with haplotype. The frequency of occurrence of a polymorphism at a particular locus provides a measure of confidence that the SNP represents a true polymorphism; this is referred to as the SNP redundancy score. In addition, true SNPs that represent divergence between homologous genes co-segregate to define a conserved haplotype. A co-segregation score based on whether a SNP position contributes to defining a haplotype provides a second independent measure of SNP confidence. The SNP redundancy score and co-segregation score together provide an effective means for estimating confidence in the validity of SNPs independently of sequence trace files (4–6).
We have combined SNP discovery software and sequence annotation within the relational database schema of autoSNPdb to enable the efficient identification of SNP and indel polymorphisms related to specific genes or traits. Here, we present the application of autoSNPdb to barley, rice and Brassica species. AutoSNPdb has a flexible interface facilitating a variety of queries. Users may search for SNPs within genes of predicted function, and through sequence identity with known genes. In addition, it is possible to add additional levels of annotation and novel queries specific to areas of interest. In the current version, we include plant cultivar information to allow the identification of SNPs that discriminate between plant cultivars.
METHODS
Data processing
Brassica, rice and barley expressed sequences were downloaded from GenBank release 159. RepeatMasker (www.repeatmasker.org) was used to identify and mask repeats prior to assembly using CAP3 (7) with the parameters –p 90, –o 50. The resulting assemblies and singleton sequences were parsed into a MySQL database. SNP discovery used the autoSNP method (4) implemented with custom Perl scripts, and the results were parsed to the database. Assemblies containing four sequences or more were examined for polymorphisms, with gaps created during the assembly process classified as indels. The minimum redundancy score defining a polymorphism was varied in proportion to the number of sequences in the assembly at the SNP position. A minimum redundancy score of 2 was required for up to seven sequences; 3 for between 8 and 11 sequences; 4 for between 12 and 19 sequences, and a minimum redundancy of 5 was required for predicting SNPs represented by 20 or more sequence reads. Each SNP was compared to all SNPs in an assembly to calculate the SNP co-segregation score, with the weighted co-segregation score calculated according to the proportion of missing data at that position in the assembly. Input sequences were annotated with cultivar type, tissue source and developmental stage where available. Consensus and singleton sequences were annotated based on sequence alignment using BLAST (8) against GenBank and UniRef90 databases. Gene Ontology (GO) annotations were derived from UniRef90 annotations. Comparative rice and Arabidopsis genome positions were derived by WU-BLAST comparison with TIGR rice pseudo-chromosomes (version 5) and TAIR Arabidopsis pseudo-chromosomes (v01222004), respectively.
Database content, access and interface
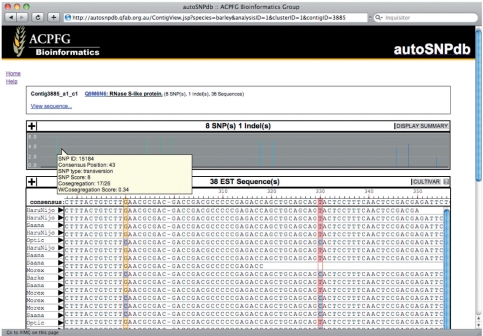
Barley, rice and Brassica sequences were downloaded from GenBank and processed through the autpSNPdb pipeline. A custom web interface allows users to query and visualize the SNP and annotation data (Figure 1). The maintenance of these data within a relational database enables numerous query options. Sequence annotations may be searched by gene keyword, sequence ID, GO term or through similarity to defined regions of the rice or Arabidopsis genome. A BLAST interface enables identification by sequence similarity. SNPs may be retrieved that differentiate between cultivars, providing a valuable resource for genetic mapping and association studies. To aid interpretation of the predicted SNP data, SNPs are viewed graphically as vertical bars, where the position of the bar along the x-axis reflects the relative position of the SNP in the consensus sequence; the height of the bar represents the SNP redundancy score; and the bar colour reflects the SNP-weighted co-segregation score. Information about each SNP is displayed by moving the cursor over the bar, while selecting a bar centres the sequence assembly at that position. The sequence assembly may be moved using the scroll bar and can be toggled between the full sequence assembly and a SNP summary. Labels to the left of the sequence may also be toggled between cultivars, GenBank accession numbers, tissue type and development stage for the respective sequences. The interface is documented with help pages and database build information.
Figure 1.
The autoSNPdb web interface displaying the sequence assembly, predicted SNPs as vertical bars and details presented in a mouse over box.
FUTURE DIRECTIONS
The autoSNPdb system was developed for flexible use and permits extension to a broad range of annotation and species. We plan to extend this system for other crops, including wheat and next-generation Roche 454 sequence data.
FUNDING
Funding for open access charge: the Australian Research Council.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Support from The National Computing Infrastructure (NCI) and the Queensland Facility for Advanced Bioinformatics (QFAB) is gratefully acknowledged. This research was supported under Australian Research Council's Linkage Projects funding scheme.
REFERENCES
- 1.Syvanen AC. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 2.Taillon-Miller P, Gu ZJ, Li Q, Hillier L, Kwok PY. Overlapping genomic sequences: a treasure trove of single-nucleotide polymorphisms. Genome Res. 1998;8:748–754. doi: 10.1101/gr.8.7.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marth GT, Korf I, Yandell MD, Yeh RT, Gu ZJ, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 4.Barker G, Batley J, O'Sullivan H, Edwards KJ, Edwards D. Redundancy based detection of sequence polymorphisms in expressed sequence tag data using autoSNP. Bioinformatics. 2003;19:421–422. doi: 10.1093/bioinformatics/btf881. [DOI] [PubMed] [Google Scholar]
- 5.Batley J, Barker G, O'Sullivan H, Edwards KJ, Edwards D. Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol. 2003;132:84–91. doi: 10.1104/pp.102.019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savage D, Batley J, Erwin T, Logan E, Love CG, Lim GAC, Mongin E, Barker G, Spangenberg GC, Edwards D. SNPServer: a real-time SNP discovery tool. Nucleic Acids Res. 2005;33:W493–W495. doi: 10.1093/nar/gki462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]