Abstract
Ovarian cancer (OC) is becoming the most common gynecological cancer in developed countries and the most lethal gynecological malignancy. It is also the fifth leading cause of all cancer-related deaths in women. The identification of diagnostic biomarkers and development of early detection techniques for OC largely depends on the understanding of the complex functionality and regulation of genes involved in this disease. Unfortunately, information about these OC genes is scattered throughout the literature and various databases making extraction of relevant functional information a complex task. To reduce this problem, we have developed a database dedicated to OC genes to support exploration of functional characterization and analysis of biological processes related to OC. The database contains general information about OC genes, enriched with the results of transcription regulation sequence analysis and with relevant text mining to provide insights into associations of the OC genes with other genes, metabolites, pathways and nuclear proteins. Overall, it enables exploration of relevant information for OC genes from multiple angles, making it a unique resource for OC and will serve as a useful complement to the existing public resources for those interested in OC genetics. Access is free for academic and non-profit users and database can be accessed at http://apps.sanbi.ac.za/ddoc/.
INTRODUCTION
In the past few years, it has become increasingly evident that ovarian cancer (OC) is a biologically complex and multigenic disease (1). Most of the genes implicated in OC (OC genes) have not been thoroughly investigated in the context of OC, especially at the gene regulation level. Understanding the regulatory mechanisms and functional operation context of the key OC genes will be useful for deciphering the impact of various molecules to the functionality of these genes, as well as effects of these genes, and thus will help to better understand different aspects of OC-gene functionality. Since information about OC genes is scattered across various resources, its integration into one resource will provide simplified way of exploring functional context of operation of OC gene, e.g. regulatory mechanisms or modes of operations. To the best of our knowledge, no database dedicated to OC genes has been published. Currently, only one database, Ovarian Kaleidoscope Database (Okdb) (2), partially addresses the needs of OC research community and is restricted to the genes expressed in the ovary of multiple species, making it more beneficial for general ovarian tissue-based research. For example, Okdb (http://ovary.stanford.edu/) originally contained information about 450 genes expressing in ovary from different species such as human, mouse, rat and bovine. In the current version, this list has now been expanded to 2788 genes. The database search using keyword ‘cancer’ retrieved only 235 genes for human and rat species combined and there is no explicit information available regarding the involvement of these genes in OC. It should be noted that there are two initiatives aimed at coordinating activities in producing resources related to cancer research, such as the International Cancer Genome Consortium—ICGC (http://www.icgc.org/) and caBIG (cancer Biomedical Informatics Grid™, http://cabig.cancer.gov/). These two intend to promote specific data formats and other conditions that should enable easier integration of cancer-related resources.
We present here the first database (Dragon Database for Exploration of Ovarian Cancer Genes, DDOC) of genes experimentally linked with OC that possess a comprehensive set of features, which allows users to explore different aspects of functionality of the OC genes indexed in DDOC and provides important information required for in-depth analysis of these genes at pathway and ontology levels. DDOC is unique for its utility to provide an opportunity to explore various gene properties that are not obtainable without complex additional analyses, such as the promoter properties and association of OC genes with other human genes, nuclear proteins, pathways and enzymes. Precompiled results of these analyses, based on promoter content and text mining, have been integrated into DDOC. We have provided ‘Batch query’ option in the search menu and user can select different types of information to be extracted from DDOC. This facility allows for downloading various selection of information from DDOC. We hope that this resource will serve as a useful complement to the existing public resources and as a good starting point for researchers and physicians interested in OC genetics, helping them get deeper insights into information about OC genes and their molecular operation modes. This information indeed will be useful for functional genomics research. DDOC is freely available at http://apps.sanbi.ac.za/ddoc/ for academic and non-profit users.
GENE SEARCH AND SELECTION CRITERIA
The gene-related information provided in the database was compiled from various repositories. Initially, a list of 900 genes was collected from sources like Cancer Gene Census (3) (http://www.sanger.ac.uk/genetics/CGP/Census/), GeneCards (4) (http://www.genecards.org/index.shtml), SymAtlas (5), OMIM (6) (online Mendelian inheritance in man, 2007) (http://www.ncbi.nlm.nih.gov/), Ovarian Kaleidoscope Database (2) (http://ovary.stanford.edu/), Entrez Gene (7) (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene) and GenAtlas (8) (http://www.genatlas.org/). Out of these 900 genes, after a thorough literature search, we compiled a final list of 379 genes that was used to populate the database. For inclusion into the database, the gene must have experimental confirmation of the differential expression in OC tissue using techniques such as: RT–PCR, immunohistochemistry, western blotting or FISH (fluorescent in situ hybridization), to name a few. Genes documented as having OC-linked SNP were also included in the database. The genes shown to have differential expression only based on microarray experiments were not included in the database.
DATABASE STATISTICS
Currently, DDOC contains a set of 379 human genes experimentally verified as involved in OC. The gene symbols, gene names and EntrezGene IDs are provided for all the genes. HGNC IDs are available for 374 genes, while Ensembl IDs are available for 370 genes. GO (9) annotations are available for 367 genes and 353 genes are indexed in eVOC (10,11). Pathways in KEGG (12) and REACTOME (13) were mapped to 211 and 50 genes, respectively. OC genes were associated with 1446 promoters. Analysis of these promoters shows that the transcription factor binding sites (TFBSs) have been predicted for 1449 TFs (Transfac IDs). Text-mining analysis involved 588 727 PubMed abstracts. The summary of statistics is provided at http://apps.sanbi.ac.za/ddoc/DDOC.pdf. DDOC will be regularly updated twice a year.
UNIQUE FEATURES OF DDOC
The identification of putative TFBSs on the promoters of OC genes could provide insights into possible regulatory characteristics of the genes. This type of information is not freely available to biologists and requires separate computational analysis. To generate these reports, we extracted 1446 promoters covering regions [−1000, +200] relative to the transcription start sites (TSS) for 371 of the 379 OC genes using the FANTOM 3 promoter set based on CAGE libraries (14,15). TFBSs were mapped to the both strands of promoter sequences using all mammalian matrix models of TFBSs contained in the TRANSFAC Professional database v.11.4 (16,17). For this purpose, we used the MatchTM program with minFP profile for thresholds of the matrix models in order to minimize false positive predictions in the predicted TFBS set (18). In a subsequent step, we extracted all mammalian TFs that are associated with the Transfac position weight matrices. In that manner, we derived an overview of hypothetical transcriptional control of the OC genes, that is, insight into TFs that are predicted to bind in the promoter regions of the genes.
One of the unique features of this database is that we have incorporated the pre-compiled results of text mining of the available literature to give an expanded view of the nuclear proteins, pathways, enzymes and mammalian genes potentially associated with each of the OC genes. As a source, we used the National Center for Biotechnology Information (NCBI) PubMed database (http://www.ncbi.nlm.nih.gov). For querying the PubMed, we created a simple tool based on the NCBI ‘Entrez Programming Utilities’. With this tool the PubMed database was queried for each gene using the following keywords:
(‘Gene Symbol’ OR ‘Gene Alias’ OR ‘Gene Alias’, etc.) AND mammal AND cancer.
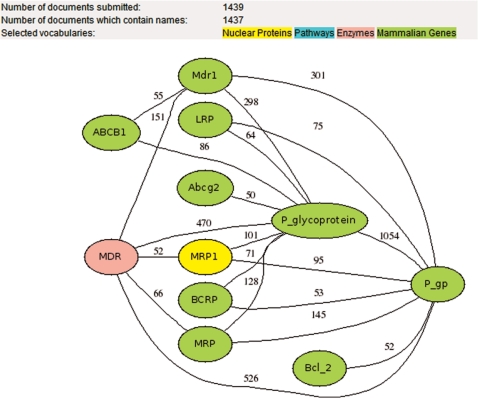
Such queries produced list of 588 727 abstracts that were analyzed by the licensed Dragon Exploration System (DES) from OrionCell (http://www.orioncell.org), that has an integrated Biomedical Text-Miner, a redeveloped tool based on the concepts from Dragon Plant Biology Explorer (19) and Dragon TF Association Miner (20). By this tool, we indexed the text document by vocabularies for nuclear proteins, pathways, enzymes and mammalian genes, and produced a database of associations that is integrated into DDOC. This integration allowed for presentation of text-mining results as lists of tables and as a graphic system of interactive networks. Figure 1 shows an example of such a network. Color-coded vocabulary entries are interconnected with weighted links representing frequency of appearance of a term and its neighbors in our abstracts list. By clicking on a node, users get relevant abstracts containing the selected term and terms representing its first neighbors (surrounding nodes in the network).
Figure 1.
A color-coded network generated from text-mining results for ABCB1 gene. Networks represents that MRP1 (a nuclear protein) appeared 95 times in the abstracts (as both have been shown to transport same substrates) with ABCB1 (P-gp), which in turn has been reviewed 470 times with medium-chain dehydrogenase/reductase (MDR) enzyme. This represents a small network of different biological entities, which have been reviewed with ABCB1 gene in the literature. This gives the user a view and understanding of varied biological interactions a gene might have in the studies published so far.
The potential uses and advantages of the database are described in the documentation section (http://apps.sanbi.ac.za/ddoc/DDOC.pdf), where various aspects of the database creation and usage have been thoroughly discussed. An example of analysis (http://apps.sanbi.ac.za/ddoc/DDOC.pdf) has been shown which should help users to understand and use different functions implemented in this database to maximize information extracted.
DISCUSSION
To date, there is no resource available, which could provide detailed information about the various aspects of the functionality and regulation of the genes known to be associated with OC. We have compiled the first database where information taken from other resources and several new analyses have been integrated into a new database resource along with details about the experimental methods used to identify the gene as a component of OC genetics. A number of components of this database are manually curated, though some parts, such as promoter analysis and text-mining components are not. The purpose of creating this database is to provide an integrated knowledgebase that allows researchers, students and clinicians to get an overview of and explore efficiently the biology of the genes involved in OC. The database provides detailed information about the homologs, regulatory mechanisms, pathways, as well as text-mining results where association of genes with other biological entities and pathways (described in literature) has been deciphered and presented as association networks rendering the incorporated information more understandable and useful. Text mining is a convenient and efficient method to summarize information from and explore the huge amount of documents in short period of time and to visualize important potential associations between different concepts in an easy to follow graphical representations. In addition to the various data-querying possibilities that our database enables, we have provided all standard facilities for users of bioinformatics databases that allow them, for example, to download data, make batch queries or take the MySQL database dump.
We hope that this database would serve as a useful resource for researchers and medical professionals who are involved in OC at any level. DDOC is aimed to serve as one-stop shop for OC research community and is created to save time and effort in order to facilitate the biological discovery process. By time, the database will be enriched by addition of new OC genes and other functionalities based on users comments.
FUTURE DIRECTIONS
DDOC reflects the information available for genes involved in OC at the period of its creation and will continue to grow both in content and functionality as more data is made available in literature. We plan to include similar information for genes differentially expressing in OC cell lines and tissue as identified by microarray and other experiments. Another line of expansion would be to incorporate the information about the drugs interacting with these genes/gene products, which will make it more useful and attractive for medical researchers and will serve a broader scientific community. Additional features that may enhance search and retrieval of DDOC information will be added in due course, as well as incorporation into ICGC and caBIG.
FUNDING
National Bioinformatics Network grants (to A.R., U.S., M.M., S.S. and V.B.B.) partially; National Research Foundation (61070 to M.M. and V.B.B.) partially; DST/NRF Research Chair (64751 to V.B.B.) and National Research Foundation (62302 to V.B.B.) partially. Funding for open access charge: National Bioinformatics Network grants.
Conflict of interest statement. V.B.B. and A.R. are partners in the OrionCell company whose product, Dragon Exploration System (DES), has been used in creation of DDOC precompiled reports. Other authors declare no conflict of interest.
ACKNOWLEDGEMENTS
M.K. has been supported by the postdoctoral fellowship from the Claude Leon Foundation, South Africa.
REFERENCES
- 1.Edlich RF, Winters KL, Lin KY. Breast cancer and ovarian cancer genetics. J. Long Term Eff. Med. Implants. 2005;15:533–545. doi: 10.1615/jlongtermeffmedimplants.v15.i5.60. [DOI] [PubMed] [Google Scholar]
- 2.Leo CP, Vitt UA, Hsueh AJ. The Ovarian Kaleidoscope database: an online resource for the ovarian research community. Endocrinology. 2000;141:3052–3054. doi: 10.1210/endo.141.9.7679. [DOI] [PubMed] [Google Scholar]
- 3.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, Ben Dor U, Esterman N, Rosen N, Peter I, et al. GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;18:1542–1543. doi: 10.1093/bioinformatics/18.11.1542. [DOI] [PubMed] [Google Scholar]
- 5.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baxevanis AD. Searching online Mendelian inheritance in man (OMIM) for information for genetic loci involved in human disease. Curr. Protoc. Hum. Genet. 2003 doi: 10.1002/0471142905.hg0913s73. Chapter 9, Unit9, 13. [DOI] [PubMed] [Google Scholar]
- 7.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frezal J. Genatlas database, genes and development defects. C. R. Acad. Sci. III. 1998;321:805–817. doi: 10.1016/s0764-4469(99)80021-3. [DOI] [PubMed] [Google Scholar]
- 9.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelso J, Visagie J, Theiler G, Christoffels A, Bardien S, Smedley D, Otgaar D, Greyling G, Jongeneel CV, McCarthy MI, et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruger A, Hofmann O, Carninci P, Hayashizaki Y, Hide W. Simplified ontologies allowing comparison of developmental mammalian gene expression. Genome Biol. 2007;8:R229. doi: 10.1186/gb-2007-8-10-r229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 15.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 16.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bajic VB, Veronika M, Veladandi PS, Meka A, Heng MW, Rajaraman K, Pan H, Swarup S. Dragon plant biology explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists. Plant Physiol. 2005;138:1914–1925. doi: 10.1104/pp.105.060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan H, Zuo L, Choudhary V, Zhang Z, Leow SH, Chong FT, Huang Y, Ong VW, Mohanty B, Tan SL, et al. Dragon TF association miner: a system for exploring transcription factor associations through text-mining. Nucleic Acids Res. 2004;32:W230–W234. doi: 10.1093/nar/gkh484. [DOI] [PMC free article] [PubMed] [Google Scholar]