Abstract
Messenger RNAs, in addition to coding for proteins, may contain regulatory elements that affect how the protein is translated. These include protein and microRNA-binding sites. Transterm (http://mRNA.otago.ac.nz/Transterm.html) is a database of regions and elements that affect translation with two major unique components. The first is integrated results of analysis of general features that affect translation (initiation, elongation, termination) for species or strains in Genbank, processed through a standard pipeline. The second is curated descriptions of experimentally determined regulatory elements that function as translational control elements in mRNAs. Transterm focuses on protein binding sites, particularly those in 3′-untranslated regions (3′-UTR). For this release the interface has been extensively updated based on user feedback. The data is now accessible by strain rather than species, for example there are 10 Escherichia coli strains (genomes) analysed separately. In addition to providing a repository of data, the database also provides tools for users to query their own mRNA sequences. Users can search sequences for Transterm or user defined regulatory elements, including protein or miRNA targets. Transterm also provides a central core of links to related resources for complementary analyses.
INTRODUCTION
Messenger RNAs are translated into proteins, directed by specific signals in the mRNA. The genetic code and codon usage may differ between species. Translation in specific organisms may also require that they make efficient use of elements around the initiation and termination codons and also use a codon bias for that organism's set of tRNAs. The preferred, often most efficient set of signals, in a particular organism can often be inferred from that most commonly used in that organism. For example, Homo sapiens has a strong bias prior to initiation codons (Kozak's consensus) (1), whereas Escherichia coli has a G/U bias following termination codons. These have been associated with efficiency of initiation and termination respectively (2,3).
In addition to this general bias reflecting overall translation, individual mRNAs may contain regulatory elements within the mRNA that affect mRNA localization, stability or translation of the associated coding region (4–6). These function most frequently in the 3′-UTR but also in 5′-UTRs or coding regions (7,8). Key known elements are protein and miRNA-binding sites (9,10). Mutations and variations in these regulatory elements have been shown experimentally to affect their function and to be underlying contributors to genetic disease (11).
DATABASE GENERATION AND CONTENT
Transterm sequences and summaries
The detail of how Transterm 2008 was generated, and software used is available on the web site. A summary including major changes in this release is presented below. Data is parsed from NCBI Genbank or NCBI Genomes entries using CDS (coding sequence) fields, and mRNA fields when available. Key regions (CDS, 5′-UTRs and 3′-UTR, Init, Term) or flanks are extracted using this CDS or mRNA information. Eight sets of data are provided for each taxonomic strain with over 40 CDS or mRNAs. The strains are identified from the TaxID (NCBI taxonomy database identifier) in the Genbank entry. Data collected can differ in experimental support and redundancy.
For ‘Genomes’ sets reducing redundancy is not done, as genomes are considered to be complete datasets, but for Genbank data redundancy is removed according to our published procedure (12). This results in redundant and non-redundant sets of regions: users choose which is appropriate to their needs. These sets of data are processed to generate summary data for each TaxID.
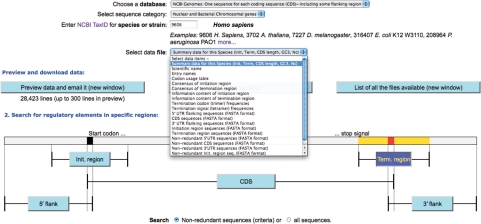
In previous releases of Transterm, data was ‘mapped up’ to the species level. With the increasing number of specific strains of a particular species now present in Genbank, we now use the strain as the taxonomic unit to collate and organize the data. For example, the 10 complete E. coli strains are processed separately, rather than combined. The sets of data are then processed as described previously to give a comprehensive set of analyses for each dataset. A view of part of the new interface is shown in Figure 1.
Figure 1.
Part of the new Transterm user interface. Users select data to analyse from four datasets, e.g. ‘NCBI Genbank—One sequence for each coding sequence entry’. A taxomic group is selected by NCBI ‘TaxId’ number (e.g. 9606), then a particular type of output (listed in Table 1) can be selected by using the pull down menu (e.g. Consensus of initiation region, Figure 2). Data selected can be for all the sequences or a non-redundant set (for H. sapiens 96 417 versus 32 763 sequences). This data can also be searched using Blast or Scan for matches.
Two files summarizing initiation codon context for two complete bacterial genomes are shown in Figure 2. This is a comparison between a section of data from the context of two eubacteria, Synechocystis PCC6803 (TaxID: 1148) and Pseudomonas aeruginosa PAO1 (TaxID: 208964) initiation codons (*.initmatrix). The upper panel shows a typical Shine-Dalgarno (SD) like pattern for a high GC% genome (for example purines at −13 to −7, whereas the lower panel PC6803 has an atypical pattern for a bacterium (less purine bias at −13 to −7, pyrimidine bias at −2, −1). Further investigation of this observation using Transterm data could utilise alternative representations of the same data, see Table 1 (Panel C) (*.initnrttbit, *.initnrttcvs), the aligned sequences themselves (*.init, *.dat) or summaries of the data (*.sum). As suggested by this data cyanobacteria have been shown to use a combination of SD-dependent and SD-independent initiation (13,14).
Figure 2.
The ‘Consensus of initiation region’ files for Synechocystis PCC6803 (NBSynePCC_2-1148.initmatrix) and Pseudomonas aeruginosa PAO1 (NBPseuaeru-208964.initmatrix). A count of the percentage of each base in each position is shown (see text for analysis). The position (Pos) in the matrix is shown above −20 to +13, the ATG is at +1 to +3. The consensus (Cons) (>65%) is shown below. For these datasets the upper sequences were 41.7% GC3 and lower 65.8% GC3. More comprehensive descriptions of the data are also available (Table 1).
Table 1.
The key output files and a brief description of the contents of each. Further descriptions are available through the online help ‘Main Transterm Datafiles’
| ClassSSN-TaxID.complete | Entries with complete CDS (have both inits + terms) |
|---|---|
| A: Lists of entries and identifiers in the redundant and non-redundant sets | |
| *.dat | Data: LOCUS, AccNo, Init [-20,+20], Term [−10,+10], Len, GC3, Nc |
| *.entry | Genbank names without descriptions |
| *.names | List of GenBank names (original input file) |
| *.text | Feature table outputs of TEXT information |
| *.TTSelected | Entries selected by reject_dups criteria |
| B: 5′-UTRs | |
| *.5UTR | 5′-UTRs/flanks, transterm format |
| *.5UTRnrtt | 5′-UTRs/flanks, non-redundant |
| *.5UTRnrtt.fa | 5′-UTRs/flanks, FASTA sequences, non-redundant |
| *.5UTR.fa | 5′-UTRs/flanks, FASTA sequences |
| C: Initiation codon context | |
| *.InitEntries | Entries in.init |
| *.init.fa | Initiation region, FASTA sequences |
| *.init | Initiation region |
| *.initmatrix | GCG consensus output for initiation region (NR) |
| *.initnrttbit | Bit scores for NR initiation region |
| *.initnrttchi | Chi scores for NR initiation region |
| *.initnrttcvs | CVS scores for NR initiation region |
| *.initnrtt.fa | Initiation region, FASTA sequences, non-redundant |
| *.initnrttver | Schneider info. scores, init. region, non-redundant |
| *.initver | Schneider information scores, init. region |
| D: CDS (coding sequences) | |
| *.CDS.fa | Full CDS entries, FASTA sequences |
| *.CDS | Full CDS entries |
| *.CDSnrtt.fa | Full CDS entries, FASTA sequences, non-redundant |
| *.CDSnrtt | Full CDS entries, non-redundant |
| E: Codon usage and bias | |
| *.cod | GCG format of codon usage |
| *.rscu | Output rscu table |
| *.sum | Summary of all the key values |
| F: Termination codon context | |
| *.TermEntries | Entries in.term |
| *.term.fa | Termination region, FASTA sequences |
| *.term | Termination region |
| *.termmatrix | GCG consensus output for termination region (NR) |
| * _termnr.summary | Count_signal of tetramer freq (readable output) |
| * _termnr.tet_tab | Termination tetramer (codon + 3′ base) frequencies |
| * _termnr.tri_tab | Termination trimer (codon) frequencies |
| *.termnrttbit | Bit scores for NR termination region |
| *.termnrttchi | Chi scores for NR termination region |
| *.termnrttcvs | CVS scores for NR termination region |
| *.termnrtt.fa | Termination region, FASTA sequences, non-redundant |
| *.termnrtt | NR version of.term, by old reject_dups criteria |
| *.termnrttver | Info. scores, term. region, non-redundant |
| *.termver | Information scores, term. region |
| G: 3′-UTRs | |
| *.3UTR.fa | 3′-UTRs/flanks, FASTA sequences |
| *.3UTR | 3′-UTRs/flanks |
| *.3UTRnrtt.fa | 3′-UTRs/flanks, FASTA sequences, non-redundant |
| *.3UTRnrtt | 3′-UTRs/flanks, non-redundant |
A list of the key classes of output files are shown in Table 1. More detail of the content of each of these files in an online help document on the website. Many of these analyses are newly available in this release.
Transterm elements
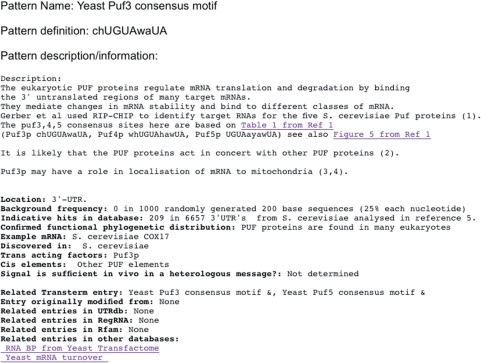
Published literature was surveyed for descriptions of new elements. New elements would be included as they become available through published literature or feedback from users. Criteria for inclusion in Transterm are that it must be experimentally verified and published in a peer reviewed journal, and that it must be sufficiently well defined to be converted into a computer readable form (regular expression, matrix, secondary structure, or discrete sequence). Some elements, e.g. the Puf3-binding site from Saccharomyces cerevisiae are currently in this form in Transterm only. The format of an example (Puf3 protein-binding site) is shown in Figure 3.
Figure 3.
An example of Transterm element description (Puf3p-binding site). Elements may be described by strings, regular expressions, matrices or RNA secondary structure rules. In this case the element is simply described as a string. Users may construct more complex descriptions of the element based on the referenced literature, for example allowing mismatches, insertions or deletions.
Where appropriate, elements reported in other databases, have been included after an independent literature review. In a similar fashion, several databases include reformatted Transterm elements (15,16). Some elements e.g. the well-studied Iron Responsive Element (IRE) are available as computer readable descriptor in several online databases, in these cases hyperlinks are provided from Transterm to allow the user to choose the most appropriate tool for analysis. Large highly structured RNA elements (e.g. riboswitches, IRESs) are not included, but are described in Rfam, ncRNA and IRESsite (17,18). The focus of Transterm is on protein-binding sites.
COMPARISON WITH OTHER TRANSLATIONAL CONTROL DATABASES
Several other databases provide some specific data, tools or services that complement those of Transterm. There is a list of resources referenced in the Transterm help online but the most relevant are summarized here. Rfam—the database of RNA families contains some cis-regulatory elements common to Transterm—these are cross-referenced. The elements are described in a different way (covariation models) and therefore are suitable for different types of analyses. RegRNA (15), UTRdb (19), Recode (20) all have related functionality but have not been updated since 2006.
Update frequency
Translational control elements are updated regularly and the sequence datasets annually.
FUNDING
Health Research Council (HRC05/195 to W.P.T., C.M.B., L.P. and R.T.P.); REANNZ and TelstraClear Capability build fund grant (CB611 to C.M.B., M.A.B.); and utilizes the NZ Biomirror and Bestgrid resources.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Thanks to users who made suggestions for improvement or gave feedback.
REFERENCES
- 1.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 2.Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cridge AG, Major LL, Mahagaonkar AA, Poole ES, Isaksson LA, Tate WP. Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2006;34:1959–1973. doi: 10.1093/nar/gkl074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol. Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Dahm R, Kiebler M, Macchi P. RNA localisation in the nervous system. Semin. Cell Dev. Biol. 2007;18:216–223. doi: 10.1016/j.semcdb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen A, Kao YF, Brown CM. Translation of the first upstream ORF in the hepatitis B virus pregenomic RNA modulates translation at the core and polymerase initiation codons. Nucleic Acids Res. 2005;33:1169–1181. doi: 10.1093/nar/gki251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 2008;18:105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JM, Ferec C, Cooper DN. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3′ UTR variants. Hum. Genet. 2006;120:301–333. doi: 10.1007/s00439-006-0218-x. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs GH, Stockwell PA, Tate WP, Brown CM. Transterm–extended search facilities and improved integration with other databases. Nucleic Acids Res. 2006;34:D37–D40. doi: 10.1093/nar/gkj159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juntarajumnong W, Incharoensakdi A, Eaton-Rye JJ. Identification of the start codon for sphS encoding the phosphate-sensing histidine kinase in Synechocystis sp. PCC 6803. Curr. Microbiol. 2007;55:142–146. doi: 10.1007/s00284-007-0057-6. [DOI] [PubMed] [Google Scholar]
- 14.Mutsuda M, Sugiura M. Translation initiation of cyanobacterial rbcS mRNAs requires the 38-kDa ribosomal protein S1 but not the Shine-Dalgarno sequence: development of a cyanobacterial in vitro translation system. J. Biol. Chem. 2006;281:38314–38321. doi: 10.1074/jbc.M604647200. [DOI] [PubMed] [Google Scholar]
- 15.Huang HY, Chien CH, Jen KH, Huang HD. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 2006;34:W429–W434. doi: 10.1093/nar/gkl333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, Kojima A, Kimura Y, Komori T, Asai K. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35:D145–D148. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mokrejs M, Vopalensky V, Kolenaty O, Masek T, Feketova Z, Sekyrova P, Skaloudova B, Kriz V, Pospisek M. IRESite: the database of experimentally verified IRES structures ( www.iresite.org) Nucleic Acids Res. 2006;34:D125–D130. doi: 10.1093/nar/gkj081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranov PV, Gurvich OL, Hammer AW, Gesteland RF, Atkins JF. Recode 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]