Abstract
Human Protein Reference Database (HPRD—http://www.hprd.org/), initially described in 2003, is a database of curated proteomic information pertaining to human proteins. We have recently added a number of new features in HPRD. These include PhosphoMotif Finder, which allows users to find the presence of over 320 experimentally verified phosphorylation motifs in proteins of interest. Another new feature is a protein distributed annotation system—Human Proteinpedia (http://www.humanproteinpedia.org/)—through which laboratories can submit their data, which is mapped onto protein entries in HPRD. Over 75 laboratories involved in proteomics research have already participated in this effort by submitting data for over 15 000 human proteins. The submitted data includes mass spectrometry and protein microarray-derived data, among other data types. Finally, HPRD is also linked to a compendium of human signaling pathways developed by our group, NetPath (http://www.netpath.org/), which currently contains annotations for several cancer and immune signaling pathways. Since the last update, more than 5500 new protein sequences have been added, making HPRD a comprehensive resource for studying the human proteome.
INTRODUCTION
Human Protein Reference Database (HPRD; http://www.hprd.org/) is a resource for experimentally derived information about the human proteome including protein–protein interactions, post-translational modifications (PTMs) and tissue expression (1–4). The contents of several proteomic databases, including HPRD, pertaining to human proteins have recently been evaluated in terms of the number of nonredundant protein–protein interactions, number of direct interactions per protein, number of proteins with disease annotation and the number of linked citations (5). The curation and annotation process in HPRD involves entry of protein data through BioBuilder, a tool developed by our group for editing and managing data through a web browser (6). We have incorporated new features, such as PhosphoMotif Finder, links to a signaling pathway resource called NetPath, Human Proteinpedia for enhanced community participation and the use of BLAST for querying mRNA/protein data. Since the last update, we have added approximately 5500 new protein sequences and corresponding information in HPRD, which now contains information on most of the human proteins including their isoforms.
‘PhosphoMotif Finder’ searches experimentally derived phosphorylation-based substrate and binding motifs
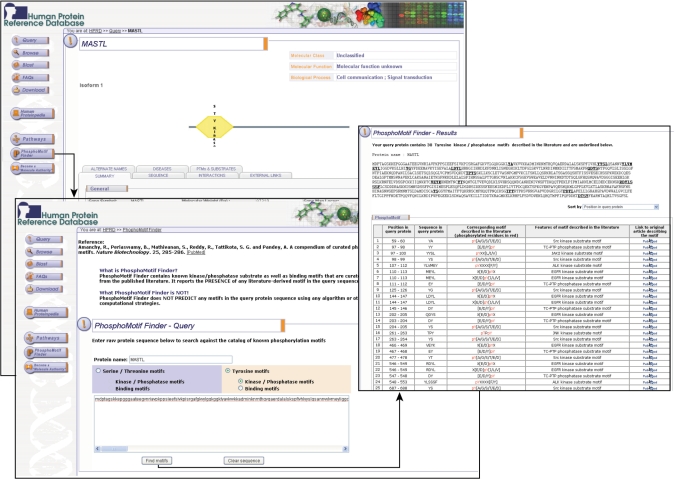
PhosphoMotif Finder contains experimentally characterized phosphorylation-based substrate and binding motifs derived from the literature (7) and has been integrated with HPRD. PhosphoMotif Finder searches across the user submitted protein sequence for the presence of any of the 320 phosphorylation-based motifs listed in the compendium. Figure 1 shows the presence of 30 known tyrosine kinase phosphorylation sites in microtubule-associated serine/threonine kinase-like protein (MASTL), which is implicated in thrombocytopenia, a blood disorder. In addition to the mapped motifs, PhosphoMotif Finder also indicates potential enzymes (i.e. kinases or phosphatases) associated with these phosphorylation motifs. PhosphoMotif Finder should also be helpful in ascertaining the novelty of any motif that is described in the literature. Finally, it can be used in designing phosphorylation motif-specific antibodies and antibody-based arrays.
Figure 1.
Display of PhosphoMotif Finder integrated into HPRD. Screen shot shows molecule page of MASTL, a hypothetical protein implicated in autosomal dominant thrombocytopenia. ‘PhosphoMotif Finder’ tab in the HPRD page leads to the utility page where the sequence of the MASTL is displayed. Users can select either serine/threonine or tyrosine motifs and submit the query by clicking ‘Find Motifs’ button. Result page displays mapped experimentally derived motifs present in sequence along with the information on position, actual sequence, experimentally derived consensus phosphorylation motifs and link to the PubMed abstracts where these motifs have been described. MASTL sequence is shown to contain 30 potential tyrosine phosphorylation sites as seen in this figure.
‘NetPath’ pathway resource
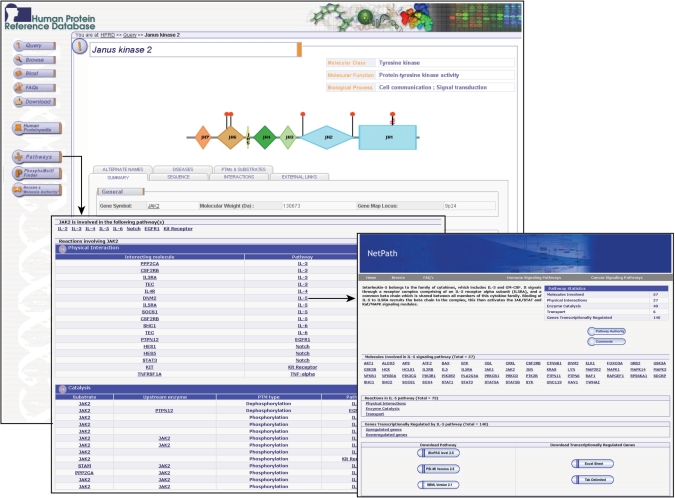
We have incorporated a compendium of human signaling pathways called NetPath (http://www.netpath.org/) through the ‘Pathways’ tab in HPRD. NetPath contains information about protein interactions, catalytic reactions and protein translocation events, which occur downstream of ligand–receptor interactions. Currently, the role of 2732 and 1793 proteins are thus annotated in the context of cancer and immune signaling pathways, respectively. We have also cataloged genes that are upregulated or downregulated at the transcriptional level under the influence of these signaling pathways. Pathway data can be downloaded in standard international data exchange formats including BioPAX Level 2.0, PSI-MI version 2.5 and SBML version 2.1. The list of transcriptionally upregulated and downregulated genes can be obtained in the form of Excel sheet and tab delimited text documents. Integration of NetPath data in HPRD will assist users in visualizing the probable role of proteins in diverse signaling networks. For example, Janus Kinase 2 (JAK2) is involved in diverse pathways including EGFR1, Kit receptor, Notch, IL-2, IL-3, IL-4, IL-5 and IL-6 signaling pathways. NetPath provides the list of physical interactions and catalysis events of JAK2 with various proteins under different signaling pathways. Each interaction or catalysis event is linked to the PubMed abstract of the original article (Figure 2).
Figure 2.
Linking to human signaling pathways from HPRD. ‘Pathways’ button in the HPRD page of JAK2 is hyperlinked to its NetPath page. It shows the list of signaling pathways in which the protein is involved along with the description of its interactors in each pathway. Each interaction or catalysis event is linked to the PubMed abstract of the original article. The pathway name is linked to the specific signaling pathway annotated in NetPath.
Annotation of proteomic information
Protein isoforms
We have included most of human protein isoforms present in the RefSeq Database (8). Currently, 25 661 protein sequences encoded by 19 433 genes have been annotated in HPRD. Phosphodiesterase 9A, cAMP response element modulator, collagen type XIII alpha1 and dystrophin are examples of proteins with the highest number of isoforms with 20, 20, 19 and 18 isoforms, respectively. However, only data pertaining to the sequence, subcellular localization, mRNA/protein expression, biological motifs and domains are currently being annotated as isoform specific whereas protein–protein interactions and enzyme–substrate relationships are annotated as common to all isoforms. This is mainly due to the general lack of experimental data for the latter.
Protein–protein interactions
Protein–protein interactions are one of the most requested components of HPRD among those who downloaded this dataset. We have added more than 5000 protein–protein interactions in HPRD since the previous update in 2006. Among the 38 167 protein–protein interactions documented in HPRD, 8958 interactions were based on yeast two-hybrid analysis alone, whereas 8827 interactions were based on in vitro and 7163 on in vivo methods. Detection of 2410 protein–protein interactions was confirmed by all three methods. Overall, in HPRD, 8710 proteins are annotated with at least one protein–protein interaction, whereas 2015 and 774 proteins have more than 5 or 10 protein–protein interactions, respectively. The 14-3-3 gamma protein has a maximum of 173 protein–protein interactions. 15 231 protein–protein interactions (Table 1) have been submitted to HPRD by the scientific community using Human Proteinpedia (9,10). Enzyme–substrate relationships determined through peptide/protein arrays is a new data type included in HPRD, as represented by the phosphorylation of Tyr 16 of RNA binding motif protein 10 by c-Src.
Table 1.
Statistics of proteomic data annotated by HPRD team and submitted to Human Proteinpedia
| Dataset | Dataset annotated by HPRD team | Data submitted through Human Proteinpedia |
|---|---|---|
| Protein–protein interactions | 38 167 | 15 231 |
| PTMs | 16 972 | 17 410 |
| Subcellular localization | 19 670 | 2906 |
| mRNA/protein expression | 65 536 | 150 368 |
PTMs and subcellular localization
HPRD currently contains information for 16 972 PTMs (Table 2) which belong to various categories with phosphorylation (10 858), dephosphorylation (3118) and glycosylation (1860) forming the majority of the annotated PTMs (Table 2). At least one enzyme responsible for PTMs has been annotated for 8960 PTMs, which resulted in the documentation of 7253 enzyme–substrate relationships. Of these, 1277 PTMs have more than one enzyme annotated. Human Proteinpedia has contributed over 17 400 PTMs, which are mainly derived from mass spectrometry studies. One or more site of subcellular localization has been annotated for 8620 proteins in HPRD with 586 of them being isoform specific. In addition to these, scientific investigators have contributed 2906 entries pertaining to subcellular localization through Human Proteinpedia.
Table 2.
Statistics of PTM data annotated among various PTM types
| PTM type | Count |
|---|---|
| Phosphorylation | 10 858 |
| Dephosphorylation | 3118 |
| Glycosylation | 1860 |
| Sumolylation | 305 |
| Acetylation | 259 |
| Methylation | 274 |
| Palmitoylation | 149 |
| Myristoylation | 43 |
| Glutathionylation | 11 |
| ADP-ribosylation | 7 |
| Others | 88 |
| Total | 16 972 |
Community participation through ‘Human Proteinpedia’
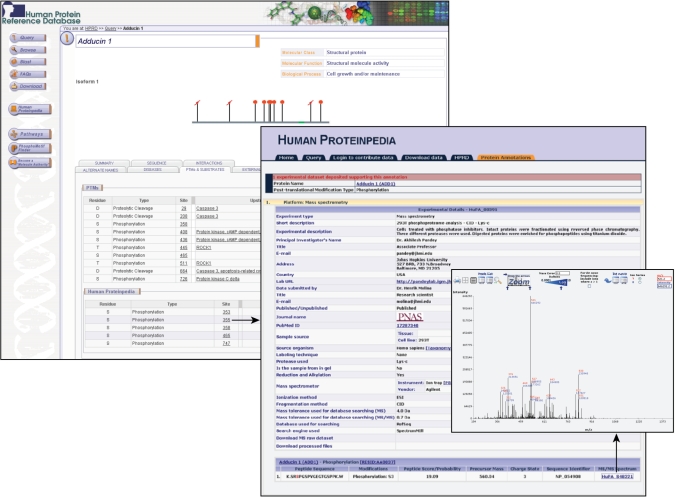
We have developed a distributed annotation system called Human Proteinpedia and incorporated in HPRD (9,10). Proteomic investigators can directly contribute protein data derived from diverse platforms including the yeast two-hybrid, mass spectrometry, peptide/protein array, immunohistochemistry, Western blot, coimmunoprecipitation and fluorescence microscopy to HPRD using Human Proteinpedia. The protein features that can be mapped to corresponding entries in HPRD include PTMs, mRNA/protein expression in tissues or cell lines, subcellular localization, enzyme–substrate relationships and protein–protein interactions. These annotations are made available for viewing in a separate box beneath the HPRD annotation (Figure 3). Each entry is also linked to experimental evidence, such as mass spectra, images of Western blots and fluorescence micrographs. Figure 3 shows five serine phosphorylation sites for Adducin 1 protein in HPRD, submitted through Human Proteinpedia. PTM sites are linked to the meta-annotation of mass spectrometry data in Human Proteinpedia database as submitted by the investigator. The corresponding MS/MS spectrum can also be viewed by following a link in the meta-annotation page.
Figure 3.
Display of PTM data in HPRD submitted through Human Proteinpedia. Adducin1 molecule page in HPRD shows five novel phosphorylation sites submitted through Human Proteinpedia. Phosphorylation sites are hyperlinked to Human Proteinpedia page with information on the investigator, laboratory and meta-annotation of mass spectrometry experiment. Corresponding MS/MS spectrum for a peptide is also displayed using spectrum viewer developed by PRIDE.
Investigators worldwide have already submitted 15 231 protein–protein interactions, 17 410 PTMs and 150 368 mRNA/protein expression to HPRD through Human Proteinpedia. Human Proteinpedia has increased quantity of the HPRD data by 2-fold in a relatively short span of time (Table 1). By involving investigators and experimentalists in the annotation of proteomic data, Human Proteinpedia has transformed HPRD into a true community database.
Usage of HPRD data by the community
Over the years, the biomedical community has provided valuable suggestions by interacting with HPRD team through ‘Comments’ and ‘Help’ buttons provided in HPRD page. More than 8000 gene comments, expert suggestions and help requests have been received and nearly 100 scientists have been designated as ‘Molecule Authorities’ based on their expertise. We hope to further increase participation by the community by implementing a microattribution system, which provides a citable credit to the investigators. Web resources that display or have made use of HPRD data include Entrez-Gene, VisANT (11) Genes2Networks (12), Cerebral (13), BioNetBuilder (14), COXPRESdb (15), STRING 7 (16) and UniHI (17). Molecular Signature Database (MSigDB) (18) used for Gene Set Enrichment Analysis of gene expression data incorporates pathway gene sets curated from HPRD. Sequence analysis tools which use HPRD data include CompariMotif (19) and SLiMFinder (20). CutDB, a database of proteolytic events (21), PepBank, a database of peptides (22) and T1Dbase, a database for type 1 diabetes research (23) are other resources that also incorporate curated proteomic data from HPRD.
CONCLUSIONS
With the inclusion of most of human protein sequences, HPRD has grown into an integrated knowledgebase for genomic and proteomic investigators. Incorporation of PhosphoMotif Finder and signaling pathways will help users to generate novel hypotheses or to point out likely molecules involved in a biological process of their interest. Further, the implementation of Human Proteinpedia has transformed HPRD into a community driven database and we hope that this trend will continue so that each and every entry is directly or indirectly verified by the individual experimentalists.
FUNDING
Funding for open access charge: Institute of Bioinformatics.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all investigators and ‘Molecule Authorities’ who have provided valuable feedback about individual entries in this database.
REFERENCES
- 1.Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat. Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 2.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database–2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathivanan S, Periaswamy B, Gandhi TK, Kandasamy K, Suresh S, Mohmood R, Ramachandra YL, Pandey A. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;7(Suppl. 5):S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro JD, Talreja N, Peri S, Vrushabendra BM, Rashmi BP, Padma N, Surendranath V, Jonnalagadda CK, Kousthub PS, Deshpande N, Shanker K, et al. BioBuilder as a database development and functional annotation platform for proteins. BMC Bioinformatics. 2004;20:5–43. doi: 10.1186/1471-2105-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nat. Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 8.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandasamy K, Keerthikumar S, Goel R, Mathivanan S, Patankar N, Shafreen B, Renuse S, Pawar H, Ramachandra YL, Acharya PK, et al. Human Proteinpedia: a unified discovery resource for proteomics research. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn701. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, et al. Human Proteinpedia enables sharing of human protein data. Nat. Biotechnol. 2008;26:164–167. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]
- 11.Hu Z, Snitkin ES, DeLisi C. VisANT: an integrative framework for networks in systems biology. Brief Bioinform. 2008;9:317–325. doi: 10.1093/bib/bbn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger SI, Posner JM, Ma’ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barsky A, Gardy JL, Hancock RE, Munzner T. Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics. 2007;23:1040–1042. doi: 10.1093/bioinformatics/btm057. [DOI] [PubMed] [Google Scholar]
- 14.Avila-Campillo I, Drew K, Lin J, Reiss DJ, Bonneau R. BioNetBuilder: automatic integration of biological networks. Bioinformatics. 2007;23:392–393. doi: 10.1093/bioinformatics/btl604. [DOI] [PubMed] [Google Scholar]
- 15.Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Res. 2008;36:D77–D82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaurasia G, Iqbal Y, Hanig C, Herzel H, Wanker EE, Futschik ME. UniHI: an entry gate to the human protein interactome. Nucleic Acids Res. 2007;35:D590–D594. doi: 10.1093/nar/gkl817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards RJ, Davey NE, Shields DC. CompariMotif: quick and easy comparisons of sequence motifs. Bioinformatics. 2008;24:1307–1309. doi: 10.1093/bioinformatics/btn105. [DOI] [PubMed] [Google Scholar]
- 20.Edwards RJ, Davey NE, Shields DC. SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins. PLoS ONE. 2007;2:e967. doi: 10.1371/journal.pone.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi Y, Eroshkin A, Gramatikova S, Gramatikoff K, Zhang Y, Smith JW, Osterman AL, Godzik A. CutDB: a proteolytic event database. Nucleic Acids Res. 2007;35:D546–D549. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shtatland T, Guettler D, Kossodo M, Pivovarov M, Weissleder R. PepBank–a database of peptides based on sequence text mining and public peptide data sources. BMC Bioinformatics. 2007;8:280. doi: 10.1186/1471-2105-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulbert EM, Smink LJ, Adlem EC, Allen JE, Burdick DB, Burren OS, Cassen VM, Cavnor CC, Dolman GE, Flamez D, et al. T1DBase: integration and presentation of complex data for type 1 diabetes research. Nucleic Acids Res. 2007;35:D742–D746. doi: 10.1093/nar/gkl933. [DOI] [PMC free article] [PubMed] [Google Scholar]