Abstract
MODBASE (http://salilab.org/modbase) is a database of annotated comparative protein structure models. The models are calculated by MODPIPE, an automated modeling pipeline that relies primarily on MODELLER for fold assignment, sequence–structure alignment, model building and model assessment (http:/salilab.org/modeller). MODBASE currently contains 5 152 695 reliable models for domains in 1 593 209 unique protein sequences; only models based on statistically significant alignments and/or models assessed to have the correct fold are included. MODBASE also allows users to calculate comparative models on demand, through an interface to the MODWEB modeling server (http://salilab.org/modweb). Other resources integrated with MODBASE include databases of multiple protein structure alignments (DBAli), structurally defined ligand binding sites (LIGBASE), predicted ligand binding sites (AnnoLyze), structurally defined binary domain interfaces (PIBASE) and annotated single nucleotide polymorphisms and somatic mutations found in human proteins (LS-SNP, LS-Mut). MODBASE models are also available through the Protein Model Portal (http://www.proteinmodelportal.org/).
INTRODUCTION
The genome sequencing efforts are providing us with complete genetic blueprints for hundreds of organisms, including humans. We are now faced with the challenge of assigning, investigating and modifying the functions of proteins encoded by these genomes. This task is generally facilitated by 3D structures of the proteins (1–3), which are best determined by experimental methods such as X-ray crystallography and NMR-spectroscopy. The number of experimentally determined structures deposited in the Protein Data Bank (PDB) more than doubled from 23 096 to 52 821 over the last 5 years (September 2008) (4). However, the number of sequences in comprehensive sequence databases, such as UniProt (5) and GenPept (6), continues to grow even more rapidly than the number of known protein structures; for example, the number of sequences in UniProt increased from 1.2 million to 6.4 million over the same period. Therefore, protein structure prediction is essential for structural characterization of sequences without experimentally determined structures.
The most accurate models are generally obtained by homology or comparative modeling (7–10), which is applicable when an experimentally determined structure related to the target sequence is available. The fraction of sequences in a genome for which comparative models can be obtained automatically varies from ∼20%–75% (11).
The process of comparative modeling usually requires the use of a number of programs to identify template structures, to generate sequence–structure alignments, to build the models and to evaluate them. In addition, various sequence and structure databases that are accessed by these programs are needed. Once an initial model is calculated, it is generally refined and ultimately analyzed in the context of many other related proteins and their functional annotations. Here, we describe MODBASE, a database of comparative protein structure models, and several associated databases and servers that facilitate modeling and analysis tasks for both expert and novice users. We highlight the improvements of MODBASE that were implemented since the last report (11), including updates in the modeling software, user interface and associated annotation tools. We also illustrate the utility of MODBASE by describing several projects depending on large model sets.
CONTENTS
Comparative modeling (MODELLER and MODPIPE)
Models in MODBASE are calculated using MODPIPE, our automated software pipeline for comparative modeling (12). It relies primarily on the various modules of MODELLER (13) for its functionality and is adapted for large-scale operation on a cluster of PCs using scripts written in PERL and Python. Sequence–structure matches are established using a variety of fold-assignment methods, including sequence–sequence (14), profile–sequence (15,16) and profile–profile alignments (16,17). Odds of finding a template structure are increased by using an E-value threshold of 1.0. By default, 10 models are calculated for each of the alignments (13). A representative model for each alignment is then chosen by ranking based on the atomic distance-dependent statistical potential DOPE (18). Finally, the fold of each model is evaluated using a composite model quality criterion that includes the coverage of the modeled sequence, sequence identity implied by the sequence–structure alignment, the fraction of gaps in the alignment, the compactness of the model and various statistical potential Z-scores (18–20). Only models that are assessed to have the correct fold were included in the final model sets.
A key feature of the pipeline is not prejudging the validity of sequence–structure relationships at the fold-assignment stage; instead, sequence–structure matches are assessed after the construction of the models and their evaluation. This approach enables a thorough exploration of fold assignments, sequence–structure alignments and conformations, with the aim of finding the model with the best evaluation score.
Comparative modeling web server (MODWEB)
MODWEB is our comparative modeling web server that is an integral module of MODBASE (http://salilab.org/modweb) (12). MODWEB accepts one or more sequences in the FASTA format and calculates their models using MODPIPE based on the best available templates from the PDB. Alternatively, MODWEB also accepts a protein structure as input, calculates a profile for each identifiable sequence homolog in the UniProt database, followed by modeling these homologs based on detectable templates in the PDB as well as the user-provided structure. Finally, MODWEB proposes a representative model based on model assessment. This module is a useful tool for measuring the impact of new structures, such as those generated by structural genomics efforts (21). The module allows us to assess the impact of a newly determined protein structure on the modeling of sequences of unknown structure. It is also used to identify new members of sequence superfamilies with at least one member of known structure. The results of MODWEB calculations are available to the users through the MODBASE interface as private datasets protected with passwords.
Pairwise and multiple structure alignments (DBAli)
DBAli (http://www.dbali.org/) stores pairwise comparisons of all structures in the PDB calculated using the program MAMMOTH (22), as well as multiple structure alignments generated by the SALIGN module of MODELLER-9 (23). DBAli contains approximately 1.7 billion pairwise comparisons and 12 732 family-based multiple structure alignments for 34 637 nonredundant protein chains out of 96 804 protein chains in the PDB. Additional information is provided by ModDom that assigns domain boundaries from structure and ModClus that allows the user to generate clusters of similar protein structures. These DBAli tools help users to analyze the protein structure space by establishing relationships between protein structures and their fragments in a flexible and dynamic manner.
Ligand binding sites (LIGBASE and AnnoLyze)
The LIGBASE module stores a list of the binding sites of known structure for approximately 230 000 ligands found in the PDB (24). The ligands include small molecules, such as metal ions, nucleotides, saccharides and peptides. Binding sites in all known structures are defined to consist of residues with at least one atom within 5 Å of any ligand atom. For each template structure, MODBASE also contains a list of putative binding sites that were predicted by the AnnoLyze program (25). The predictions are based on inheriting an actual binding site from any related known structure if at least 75% of the binding site residues are within 4 Å of the template residues in a global superposition of the two structures in DBALI and if at least 75% of the binding site residue types are invariant. In addition, the putative ligand binding sites in the models are then mapped via the target–template alignments. The putative ligand binding sites are stored as SITE records and the binding site membership frequency per residue is indicated in the B-factor column of the model coordinate files. Sixty-five percent of MODBASE models have at least one predicted binding site.
Protein interactions (PIBASE)
PIBASE (http://pibase.janelia.org, http://salilab.org/pibase) is a comprehensive database of structurally defined protein interfaces (26). It is composed of binary interfaces between pairs of chains or domains extracted from structures in the PDB and the Probable Quaternary Structure server PQS using domain assignments from the Structural Classification of Proteins and CATH fold classification systems. PIBASE currently contains 269 821 SCOP, 269 438 CATH, and 216 739 chain binary interfaces. A diverse set of geometrical, physiochemical and topological properties are calculated for each complex, its domains, interfaces and binding sites. The database is accessible through the web server and can also be installed locally. The software used to build PIBASE is available for download under an open-source license.
PIBASE is a convenient resource for structural information on protein–protein interactions and is easily integrated with other databases. It is currently used by the AnnoLyze annotation program (27) and the LS-SNP annotation system (28). The complexes stored in PIBASE can also be used as templates to predict the composition and structure of protein complexes using comparative modeling followed by an assessment of the modeled interface (29). This approach was applied to predict host–pathogen interactions for 10 ‘neglected’ human pathogens (30).
Single nucleotide polymorphisms and somatic mutations (LS-SNP and LS-Mut)
LS-SNP [http://karchinlab.org/LS-SNP, http://salilab.org/LS-SNP (28)] and LS-Mut [http://karchinlab.org/LS-Mut, (31,32)] are collections of annotated DNA sequence variants in protein-coding exons that result in an amino acid residue-type substitution. These resources focus on inherited genetic variants and tumor-derived somatic mutations, respectively. For LS-SNP, genomic locations of the variants are taken from the dbSNP database (33) and are mapped onto as many human proteins in the UniProt database (34) as possible. The mapping is achieved via a collection of protein-to-mRNA and mRNA-to-genome alignments produced with the Known Genes algorithm (35). For LS-Mut, somatic mutation data from tumor sequencing projects are used, consisting of transcript identifiers from RefSeq, CCDS and Ensembl (36,37), codon positions and amino acid residue-type substitutions. Our software then maps the mutations onto translated protein sequences. LS-Mut currently includes mutations from 24 advanced pancreatic cancers and 22 glioblastoma multiforme (brain) tumors. For both LS-SNP and LS-Mut, human protein sequences are aligned with homologous proteins of known structure from PDB, to build comparative protein structure models using MODPIPE. Models are constructed for all significant alignments covering a distinct region of protein sequence (E-value cutoff 0.0001). UCSF Chimera (38) is used to visualize the location of the residue substitutions on the model. We use our software and DSSP (39) to identify secondary structure elements and relative solvent accessibility of the residue positions. Putative protein and small ligand binding sites on the models are annotated with PIBASE and the LIGBASE module of MODBASE, respectively, to infer which SNPs or somatic mutations may destabilize protein quaternary structure or interfere with small molecule ligand binding.
MODBASE MODEL SETS
Models in MODBASE are organized into a number of datasets. The largest dataset contains models of all sequences in the UniProt database that are detectably related to at least one known structure in the PDB from July 2005. Because of the rapid growth of the public sequence databases, we now concentrate our efforts on adding datasets that are useful for specific projects, rather than attempt to model all known protein sequences with detectable template structures. Currently, MODBASE includes datasets of nine archaeal genomes, 13 bacterial genomes and 18 eukaryotic genomes (Table 1). Together with other project-oriented datasets, MODBASE currently contains 5 152 695 models from domains in 1 593 209 unique sequences. Next, we illustrate the utility of MODBASE by outlining several recent projects.
Table 1.
MODBASE datasets
| Dataset/Project | Taxonomy ID | No. of Transcripts | No. of Sequences modeled | No. of Models | Sequence source |
|---|---|---|---|---|---|
| Genomes (*genomes for the TDI) | |||||
| Archaea | |||||
| Archaeoglobus fulgidus | 2234 | 2409 | 1794 | 3980 | NCBI |
| Methanococcus jannaschii | 2190 | 1785 | 1480 | 1707 | NCBI |
| Nanoarchaeum equitans | 160 232 | 536 | 447 | 496 | NCBI |
| Picrophilus torridus | 82 076 | 1535 | 1260 | 2902 | NCBI |
| Pyrobaculum aerophilum | 13 773 | 2600 | 1566 | 3497 | NCBI |
| Pyrococcus furiosus | 2261 | 2113 | 1524 | 3373 | NCBI |
| Sulfolobus solfataricus | 2287 | 2922 | 2006 | 4451 | NCBI |
| Thermoplasma volcanium | 50 339 | 1497 | 1204 | 2806 | NCBI |
| Thermoplasma acidophilum | 1480 | 1220 | 2801 | NCBI | |
| Bacteria | |||||
| Bacillus subtilis | 1423 | 4105 | 3374 | 9245 | NCBI |
| Burkholderia mallei | 13 373 | 4798 | 3910 | 23 219 | NCBI |
| Clostridium tetani | 1513 | 2413 | 2158 | 5864 | NCBI |
| Escherichia coli | 562 | 4206 | 3150 | 5994 | NCBI |
| Mycobacterium leprae* | 1769 | 1605 | 1178 | 2493 | OrthoMCL-DB |
| Mycobacterium tuberculosis* | 1773 | 3991 | 2808 | 5913 | TubercuList |
| Mycoplasma pneumoniae | 2104 | 687 | 426 | 857 | NCBI |
| Pseudomonas aeruginosa | 287 | 5559 | 3806 | 9222 | NCBI |
| Rickettsia prowazekii | 782 | 835 | 754 | 2136 | NCBI |
| Staphylococcus aureus MRSA252 | 282 458 | 2635 | 1184 | 3161 | NCBI |
| Streptococcus pyogenes | 1314 | 1691 | 1440 | 3984 | NCBI |
| Wolbachia* | 953 | 805 | 621 | 1873 | TIGR |
| Yersinia pestis | 632 | 3882 | 3215 | 8371 | NCBI |
| Eukaryota | |||||
| Arabidopsis thaliana | 3702 | 30 707 | 23 807 | 70 494 | ENSEMBL |
| Brugia malayi* | 6279 | 11 397 | 7850 | 23 219 | TIGR |
| Caenorhabditis elegans | 6239 | 22 698 | 18 996 | 52 235 | NCBI |
| Canis familiaris | 9615 | 30 264 | 22 614 | 65 617 | ENSEMBL |
| Cryptosporidium hominis* | 237 895 | 3886 | 1614 | 3287 | CryptoDB |
| Cryptosporidium parvum* | 5807 | 3806 | 1918 | 3969 | CryptoDB |
| Danio rerio | Calculation in progress | ENSEMBL | |||
| Drosophila melanogaster | 7227 | 17 104 | 9381 | 24 683 | NCBI |
| H.sapiens* | 9606 | 32 010 | 21 270 | 51 084 | OrthoMCL-DB |
| Leishmania major* | 5664 | 8274 | 3975 | 8285 | GeneDB |
| Mus musculus | 10 090 | 30 133 | 25 338 | 70 783 | NCBI |
| Pan troglodytes | Calculation in progress | ENSEMBL | |||
| Plasmodium falciparum* | 5833 | 5363 | 2599 | 5053 | PlasmoDB |
| Plasmodium vivax* | 5855 | 5342 | 2359 | 4670 | PlasmoDB |
| Rattus norvegicus | Calculation in progress | ENSEMBL | |||
| Saccharomyces cerevisiae | 4932 | 6600 | 3035 | 5543 | NCBI |
| Schistosoma mansoni* | 6183 | 25 304 | 8576 | 26 076 | GeneDB |
| Toxoplasma gondii* | 5811 | 7793 | 1530 | 3064 | ToxoDB |
| Trypanosoma brucei* | 5691 | 9210 | 3900 | 8054 | GeneDB |
| Trypanosoma cruzi* | 5693 | 19 607 | 7390 | 14 858 | GeneDB |
| Xenopus laevis | 8355 | 27 952 | 25 457 | 69 191 | NCBI |
| Selected projects | |||||
| CSMP datasets | 195 235 | 184 139 | 690 255 | GENPEPT NR | |
| NYSGXRC datasets | 553 537 | 493 672 | 1 415 237 | GENPEPT NR | |
| Enzyme Specificity Project | 15 833 | 10 875 | 183 591 | SFLD/NR | |
| ABC Transporter | 152 | 85 | 85 | ||
| GPCR | 11 586 | 11 551 | 24 272 | ||
| UNIPROT Datasets 2005 | 1 742 816 | 1 025 196 | 2 146 830 | UNIPROT | |
| Total (including other datasets) | 2 608 987 | 1 593 209 | 5 152 695 |
Structural genomics of the enolase and amidohydrolase superfamilies
Comparative models of enzymes in the amidohydrolase and enolase superfamilies have contributed to studying their substrate specificity by the Enzyme Specificity Consortium (ENSPEC) as well as selecting targets for a structural genomics effort by the New York SGX Research Center for Structural Genomics (NYSGXRC). In particular, we selected 535 target proteins from 130 genomes for high-throughput structure determination by X-ray crystallography, resulting in 61 unique structures thus far. Both template-based modeling and sequence-based modeling were essential in identifying suitable targets.
Structural genomics of membrane proteins
Comparative modeling was also applied to inform target selection for the structural genomics of membrane proteins as part of the Center for Structures of Membrane Proteins (CSMP) at UCSF (40). The goal of CSMP is to express, purify and determine the structures of representative members of integral membrane protein classes. MODBASE models were combined with an interactive web-based target selection tool to facilitate selection of biologically interesting targets with little or no structural data available. In addition, template-based modeling in MODWEB is being used to calculate how many sequences can be modeled based on newly determined CSMP structures.
ABC Transporters
ABC transporters are a large and diverse set of integral membrane proteins that couple the action of ATP binding, hydrolysis and release to substrate transport across a cellular membrane (41). Mutations in 13 of the 48 human ABC transporters are associated with monogenic human disease phenotypes (42). Additional variants are being identified in hundreds of individuals by the Pharmacogenomics of Membrane Transporters (PMT) consortium at UCSF (43). To annotate these variants, we modeled nucleotide binding and membrane spanning domains with detectably related template structures in all human ABC transporters. The dataset also includes models of sequences with disease-associated and polymorphic nonsynonymous SNPs found in the nucleotide binding domains. Finally, the incomplete or unsatisfactory modeling coverage was used to suggest specific targets for a structural genomics effort on ABC transporters by CSMP.
Human caspases
Caspases are cysteine proteases involved in multiple apoptotic pathways. An experimental approach was recently developed to identify caspase substrates by biotinylating natural protein N-termini and selecting protein fragments containing unblocked α-amines characteristically generated upon proteolytic cleavage (44). Likely high accuracy models of protein substrates prior to cleavage were identified in the MODBASE human genome datasets and analysis of the structural properties of the cleavage sites was performed. While these sites often appeared in disordered, solvent accessible regions of the substrate as expected (45), a surprising number were found in α-helices and partially inaccessible regions, information which can now be incorporated into new algorithms for predicting additional caspase substrates.
Binding sites and ligands for the tropical disease initiative
Open source drug discovery is an alternative avenue to conventional patent-based drug development, illustrated by the proposed Tropical Disease Initiative (TDI) (http://tropicaldisease.org) (46). Open source drug discovery involves a decentralized, web-based and community-wide collaboration, in which scientists from laboratories, universities, institutes and corporations volunteer to work together for a common cause. To contribute to this effort, we calculated comparative protein structure models for 10 genomes of organisms that cause ‘neglected’ tropical diseases (Table 1). We followed up by predicting binding sites for known drugs using the AnnoLyze program (25). These predictions may be used as a starting point for experimentally testing the biological functions of the target proteins and potentially even as leads for drug discovery.
Host–pathogen protein interactions for TDI
Pathogens have evolved numerous strategies to infect their hosts, while hosts have evolved immune responses and other defenses to these foreign challenges. The vast majority of host–pathogen interactions involve protein–protein recognition, yet our current understanding of these interactions is limited. We developed and applied a computational whole-genome protocol that generates testable predictions of host–pathogen protein interactions (30) (http://salilab.org/hostpathogen). The protocol first scans the host and pathogen genomes for proteins with similarity to known protein complexes, then assesses these putative interactions, using structure if available, and, finally, filters the remaining interactions using biological context, such as the stage-specific expression of pathogen proteins and tissue expression of host proteins. The technique was applied to 10 pathogens, using their MODBASE model datasets. Several specific predictions have been made that warrant experimental follow-up, including interactions from previously characterized mechanisms, such as cytoadhesion and protease inhibition, as well as suspected interactions in hypothesized networks, such as apoptotic pathways.
G-Protein Coupled receptors
G-protein coupled receptors (GPCR) are a large family of pharmacologically important transmembrane receptors that are involved in the recognition of a wide variety of extra-cellular ligands. It has been estimated that this family of proteins is the target for about half of all currently marketed drugs. Atomic structures are known for only three sub-families of GPCRs, including light-sensitive rhodopsins, β1 and β2 adrenergic receptors that all belong to the Class A Rhodopsin-like family (GPCRDB nomenclature). The GPCR dataset in MODBASE consists of models for approximately 12 000 UniProt sequences that are related to one of these structures. The models span several sub-families of the Class A Rhodopsin-like family, including aminergic, peptide, hormone, opsin, olfactory and nucleotide receptors. These models are used for ligand docking and virtual screening computations by DOCK (47).
ACCESS AND INTERFACE
The main access to MODBASE is through its web interface at http://salilab.org/modbase, by querying with Uniprot and GI identifiers, gene names, annotation keywords, PDB codes, datasets, organisms, sequence similarity to the modeled sequences (BLAST) and model-specific criteria such as model reliability, model size and target–template sequence identity. Additionally, it is possible to retrieve coordinate files, alignment files and ligand-binding information in text files. Select genome datasets are also available from our ftp server (ftp://salilab.org/databases/modbase/projects).
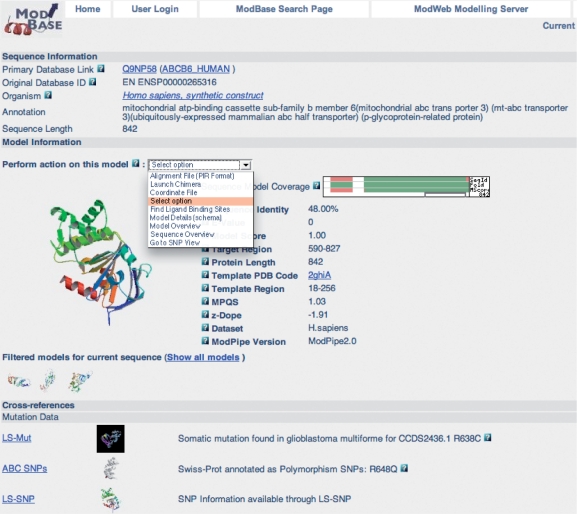
The output of a search is displayed on pages with varying amounts of information about the modeled sequences, template structures, alignments and functional annotations. An example of the output from a search resulting in one model is shown in Figure 1. A ribbon diagram of the model with the highest target–template sequence identity is displayed by default, together with details of the modeling calculation. Ribbon thumbprints of additional models for this sequence link to corresponding pages with more information. The ribbon diagrams are generated on the fly using Molscript (48) and Raster3D (49). A pull-down menu provides links to additional functionality: the ligand-binding module, the SNP module, retrieval of coordinate and alignment files, as well as molecular visualization by Chimera that allows the user to display template and model coordinates together with their alignment. If mutation information is available for a protein sequence, links to the details are provided in the cross-references section. Additionally, cross-references to various other databases, including PDB, UniProt, SwissProt/TrEMBL, PubMed and the UCSC Genome Browser, are given. Other MODBASE pages provide overviews of more than one sequence or structure. All MODBASE pages are interconnected to facilitate easy navigation between different views.
Figure 1.
MODBASE Model Details page (Example Q9NP58 from the human genome dataset): this page provides links to all models for this specific sequence. A ribbon diagram of the primary model, database annotations and modeling details are displayed. Links to additional models for different target regions or models from other datasets are displayed as thumbprints. The pull-down menu provides access to alternative MODBASE views and other types of information (if available), such as data about mutations and putative ligand binding sites. The cross-references section contains links to relevant internal and external databases. For this particular sequence, mutation data are available from LS-Mut, LS-SNP and ABC SNPs.
Access through external databases
MODBASE models in academic and public datasets are directly accessible from several other databases, including the SwissProt/TrEMBL sequence pages, UniProt, PIR's iProClass, EBI's InterPro, the UCSC Genome Browser and PubMed (LinkOut). Importantly, MODBASE models are also accessible through the Protein Model Portal (http://proteinmodelportal.org), a module of the Protein Structure Initiative Knowledgebase (PSI KB). The Model Portal has the potential to become the single entry point for users interested in experimentally determined or computationally predicted models. For a user query, the portal will interrogate participating source model databases and modeling servers to provide a comprehensive view of all available models of the query sequence.
FUTURE DIRECTIONS
MODBASE will grow by adding models calculated on demand by external users (using MODWEB) as well as our own calculations of model datasets that are needed for our research projects (using MODPIPE, MODWEB or MODELLER). These updates will reflect improvements in the methods and software used for calculating the models as well as the new template structures in the PDB and new sequences in UniProt. In the future, we expect that most of the users will access MODBASE models through the Protein Model Portal.
CITATION
Users of MODBASE are requested to cite this article in their publications.
FUNDING
National Institutes of Health (R01 GM54762, U54 GM074945, U54 GM074929, U01 GM61390, P01 GM71790 to A.S., GM08284 to D.E., NSF EF 0626651); the Sandler Family Supporting Foundation (to A.S.); Susan G. Komen Foundation (KG080137 to R.K.); Spanish Ministerio de Educación y Ciencia (BIO2007/66670 to M.A.M-R). Funding for open access charge: U54 GM074945.
ACKNOWLEDGEMENTS
We are grateful to Tom Ferrin, Daniel Greenblatt, Conrad Huang and Tom Goddard for CHIMERA and contributing to the MODBASE/CHIMERA interface. For linking to MODBASE from their databases, we thank Torsten Schwede (Protein Model Portal), David Haussler and Jim Kent (UCSC Genome Browser), Amos Bairoch (SwissProt/TrEMBL), Rolf Apweiler (InterPro), Patsy Babbitt (SFLD) and Cathy Wu (PIR/iProClass). We are also grateful for computing hardware gifts from Mike Homer, Ron Conway, NetApp, IBM, Hewlett Packard and Intel.
REFERENCES
- 1.Domingues FS, Koppensteiner WA, Sippl MJ. The role of protein structure in genomics. FEBS Lett. 2000;476:98–102. doi: 10.1016/s0014-5793(00)01678-1. [DOI] [PubMed] [Google Scholar]
- 2.Brenner SE, Levitt M. Expectations from structural genomics. Protein Sci. 2000;9:197–200. doi: 10.1110/ps.9.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skolnick J, Fetrow JS, Kolinski A. Structural genomics and its importance for gene function analysis. Nat. Biotechnol. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- 4.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 8.Wallner B, Elofsson A. All are not equal: a benchmark of different homology modeling programs. Protein Sci. 2005;14:1315–1327. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillisch A, Pineda LF, Hilgenfeld R. Utility of homology models in the drug discovery process. Drug Discov. Today. 2004;9:659–669. doi: 10.1016/S1359-6446(04)03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Curr. Protocols Protein Sci./editorial board, John E. Coligan … et al. 2007;Chapter 2 doi: 10.1002/0471140864.ps0209s50. Unit 29. [DOI] [PubMed] [Google Scholar]
- 11.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eswar N, John B, Mirkovic N, Fiser A, Ilyin VA, Pieper U, Stuart AC, Marti-Renom MA, Madhusudhan MS, Yerkovich B, et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 14.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Protocols Bioinformatics/editoral board, Andreas D. Baxevanis … et al. 2006;Chapter 5 doi: 10.1002/0471250953.bi0506s15. Unit 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marti-Renom MA, Madhusudhan MS, Sali A. Alignment of protein sequences by their profiles. Protein Sci. 2004;13:1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eramian D, Shen MY, Devos D, Melo F, Sali A, Marti-Renom MA. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melo F, Sanchez R, Sali A. Statistical potentials for fold assessment. Protein Sci. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chance MR, Fiser A, Sali A, Pieper U, Eswar N, Xu G, Fajardo JE, Radhakannan T, Marinkovic N. High-throughput computational and experimental techniques in structural genomics. Genome Res. 2004;14:2145–2154. doi: 10.1101/gr.2537904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marti-Renom MA, Ilyin VA, Sali A. DBAli: a database of protein structure alignments. Bioinformatics. 2001;17:746–747. doi: 10.1093/bioinformatics/17.8.746. [DOI] [PubMed] [Google Scholar]
- 24.Stuart AC, Ilyin VA, Sali A. LigBase: a database of families of aligned ligand binding sites in known protein sequences and structures. Bioinformatics. 2002;18:200–201. doi: 10.1093/bioinformatics/18.1.200. [DOI] [PubMed] [Google Scholar]
- 25.Marti-Renom MA, Rossi A, Al-Shahrour F, Davis FP, Pieper U, Dopazo J, Sali A. The AnnoLite and AnnoLyze programs for comparative annotation of protein structures. BMC Bioinformatics. 2007;8(Suppl. 4):S4. doi: 10.1186/1471-2105-8-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 27.Marti-Renom MA, Pieper U, Madhusudhan MS, Rossi A, Eswar N, Davis FP, Al-Shahrour F, Dopazo J, Sali A. DBAli tools: mining the protein structure space. Nucleic Acids Res. 2007;35:D393–D397. doi: 10.1093/nar/gkm236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 29.Davis FP, Braberg H, Shen MY, Pieper U, Sali A, Madhusudhan MS. Protein complex compositions predicted by structural similarity. Nucleic Acids Res. 2006;34:2943–2952. doi: 10.1093/nar/gkl353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis FP, Barkan DT, Eswar N, McKerrow JH, Sali A. Host pathogen protein interactions predicted by comparative modeling. Protein Sci. 2007;16:2585–2596. doi: 10.1110/ps.073228407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human Glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 36.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 40.Li M, Hays FA, Roe-Zurz Z, Vuong L, Kelly L, Robbins R, Ho CM, Pieper U, O'C;onnell J, Miercke LJ, et al. Eukaryotic Integral Membrane Protein Production For Structural Genomics. J. Mol. Biol., in press. 2008 doi: 10.1016/j.jmb.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 42.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc. Natl Acad. Sci. USA. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- 46.Maurer SM, Rai A, Sali A. Finding cures for tropical diseases: is open source an answer? PLoS Med. 2004;1:e56. doi: 10.1371/journal.pmed.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plorts of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 49.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 50.Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen F, Mackey AJ, Stoeckert C.J., Jr., Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole ST. Learning from the genome sequence of Mycobacterium tuberculosis H37Rv. FEBS Lett. 1999;452:7–10. doi: 10.1016/s0014-5793(99)00536-0. [DOI] [PubMed] [Google Scholar]
- 53.Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, Rhodes P, Wang S, He CZ, Su Y, et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34:D419–D422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pegg SC, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]