Abstract
A dramatic exception to the general pattern of single-copy genes in bacterial and archaeal genomes is the presence of 1–15 copies of each ribosomal RNA encoding gene. The original version of the Ribosomal RNA Database (rrnDB) cataloged estimates of the number of 16S rRNA-encoding genes; the database now includes the number of genes encoding each of the rRNAs (5S, 16S and 23S), an internally transcribed spacer region, and the number of tRNA genes. The rrnDB has been used largely by microbiologists to predict the relative rate at which microbial populations respond to favorable growth conditions, and to interpret 16S rRNA-based surveys of microbial communities. To expand the functionality of the rrnDB (http://ribosome.mmg.msu.edu/rrndb/index.php), the search engine has been redesigned to allow database searches based on 16S rRNA gene copy number, specific organisms or taxonomic subsets of organisms. The revamped database also computes average gene copy numbers for any collection of entries selected. Curation tools now permit rapid updates, resulting in an expansion of the database to include data for 785 bacterial and 69 archaeal strains. The rrnDB continues to serve as the authoritative, curated source that documents the phylogenetic distribution of rRNA and tRNA genes in microbial genomes.
INTRODUCTION
Ribosomes play a central role in every form of life by catalyzing the mRNA-dependent synthesis of proteins from amino acids. Crystal structures of this ribonucleoprotein complex reveal a catalytic center that consists primarily of ribosomal RNAs (rRNA) (1). Due to the conserved function of ribosomes, the 3D structure of the rRNAs is highly constrained, with regions of strong primary sequence conservation interspersed with variable regions. These characteristics make the molecule ideal for establishing the evolutionary relatedness of organisms, and for culture-independent molecular surveys of microbial communities.
As the applications of phylogenetic analyses and molecular surveys have expanded in microbiology, databases of aligned rRNA gene sequences including SILVA (2), the Ribosomal Database Project (3) and Greengenes (4) were created to assist in sequence analysis. However, these databases do not include information about a crucial characteristic of rRNA genes that influences molecular surveys: the number of rRNA genes per genome.
Genes encoding the 16S rRNA (rrs), 23S rRNA (rrl) and 5S rRNA (rrf) are typically arranged into an operon (rrn operon), with an internally transcribed spacer (ITS) between the 16S and 23S rRNA genes that is also used to discriminate amongst closely related organisms. The number of rrn operons ranges from one to 15 per genome. This redundancy must be considered in studies that measure the abundance of rRNA genes, especially techniques such as terminal restriction fragment length polymorphism (tRFLP), denaturing gradient gel electrophoresis (DGGE) and quantitative PCR (5). Due to redundancy of the rRNA genes in some organisms, the measured abundance of an rRNA gene might be attributed to few organisms with many rRNA genes or many organisms with few rRNA genes.
An additional benefit of knowing the rrn copy number of an organism is derived from the positive correlation between the number of rRNA genes in an organism's genome and the capacity of that organism to respond to favorable growth conditions (6, 7). This relationship suggests that the number of rRNA genes copy number reveals the life history of an organism, where organisms with few rrn operons tend to be slow growing organisms that can utilize resources efficiently, while those with many rrn operons grow more rapidly in response to favorable growth conditions but with less efficient use of resources. In addition, microbes with few rrn operons tend to be oligotrophic, i.e. capable of growth in low-nutrient environments (8, 9).
The value in linking life histories to the number of rRNA genes prompted the compilation of available information on 16S rRNA-encoding genes in the first iteration of the rrnDB (10). Microbiologists used the database as a reference for estimation of an organism's capacity to respond to favorable growth conditions, and to interpret abundance data from molecular surveys. In particular, the database has become critical for studies using quantitative PCR to enumerate bacteria and archaea in the environment (11).
Increasing use of the rrnDB and expansion of the regions of the rrn operon that are now included in molecular surveys motivated its redesign and expansion. The rrnDB is now based on a relational database that includes information on redundancy of all rRNA genes (rrs, rrl, rrf) and the number of ITS regions. The number of transfer RNA (tRNA) genes per genome has also been added to the database because it varies with rrs copy number. Users now have access to expanded queries that include dynamic calculation of average gene copy numbers for group of organisms selected, and additional search and sorting features. Curatorial tools have also been added to facilitate updating. As a result, the number of bacterial and archaeal strains included in the rrnDB has more than doubled, with 785 bacterial and 69 archaeal strains, and is now being updated regularly.
DATABASE DESCRIPTION
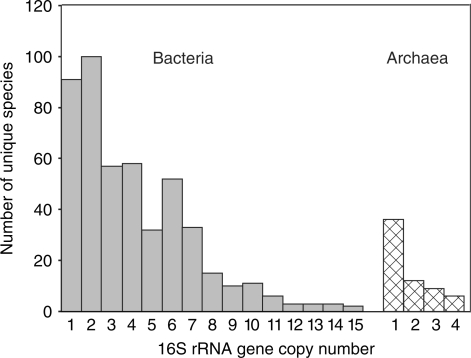
The redesigned website and database are accessible on the WWW at http://ribosome.mmg.msu.edu/rrndb. Entries in the database not only consist primarily of data from sequenced genomes, but also include data from strains in which the number of rrn genes has been determined through other methods. Among unique species in the database, 40% of bacteria have either one or two copies of the 16S rRNA gene (Figure 1). Bacterial species with eight or more 16S rRNA genes make up 11% of the unique entries, and they consist of bacteria in the phylum Firmicutes or the class γ-Proteobacteria; there is a single β-Proteobacteria, Chromobacterium violaceum, currently in this grouping. Two organisms with 15 rrn operons are know—Clostridium paradoxum and Photobacterium profundum. The range of rrn genes in archaea is smaller, with one to four copies of the 16S rRNA gene. More than half (57%) of sequenced archaeal genomes have a single copy of each of the rrn genes. Archaea with two or more 16S rRNA genes are all from the phylum Euryarchaeota.
Figure 1.
The number of 16S rRNA genes in bacterial and archaeal genomes. The analysis was performed on 476 bacterial species (gray bars) and 63 archaeal species (checkered bars).
There are multiple ways to access entries in the database: users can browse through the entire database; ‘search by keyword’, which is based on an organism's name or strain designation or simply by the number of 16S rRNA genes; ‘search by taxonomy’ allows users to select entries within a particular taxonomic level from a pull-down menu; or through combinations of these searches. The new search features also include dynamic calculation of average gene copy number for any subset of organisms selected in a search.
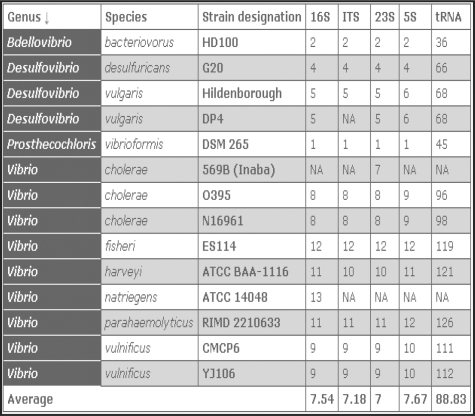
Results from database searches are presented in a table that appears below the search form (Figure 2). For each entry, the table presents the genus, species, strain designation and copy numbers for 16S, 23S, 5S rRNA, tRNA genes and the ITS. The majority of the entries will have the same number of rrs, rrl, rrf and the ITS, which might be expected because ribosomes are made up of a single transcript from each gene. However, 23.6% of genomic bacterial entries have unequal copies of the rRNA genes, due mainly to additional copies of the 5S rRNA gene. Other variations include Borrelia sp. which maintains two copies of the 23S–5S rRNA genes and one copy of the 16S rRNA gene encoded separately on the genome, and Thermobispora bispora which has four copies of 16S rRNA gene, three copies of 23S rRNA gene and only two copies of 5S rRNA gene (12, 13).
Figure 2.
A screenshot of the result table from the rrnDB using ‘search by keyword’ for ‘Vibrio’. The average gene copy number is presented at the end of the table. The dark gray highlighted column indicates that the table is sorted according to the genus name. NA indicates that information for the particular gene is ‘not available’.
For convenience, the entries in the result table can be sorted according to any column by clicking on the column heading (Figure 2). A major addition to the database is the capacity for dynamic calculation of the average copy number for any collection of organisms listed in the result table: the arithmetic average is presented for each gene at the end of the table. This feature will be particularly useful for researchers using quantitative PCR to enumerate the abundance of a specific group of organisms. The number of 16S rRNA genes is typically constant among different strains of the same species, but in ∼5% of bacterial species in the database, the number varies by one for different strains. For closely related species, the number of rrn genes per genome is often similar, but it is not entirely consistent with phylogenetic relationships. For instance, strains of both γ-proteobacteria and clostridia maintain from 1 to 15 copies.
Detailed information for each organism in the table can be viewed by clicking on the strain designation. Information will be presented below the results table. It includes organismal taxonomy, copy number for each gene and ITS, accession number for the gene entries in Genbank, genome size, genome accession number in Genbank, reference link to Entrez Pubmed and a comment section. For organisms with multiple chromosomes, the allocation of both rRNA and tRNA genes into each chromosome is described. The comment section also specifies the method used to determine the number of rRNA genes for entries not from genomic sequences. The two most common alternative methods for estimating the number of rRNA genes are Southern hybridization with rRNA gene or ITS-specific probes, and digestion with the restriction endonuclease ICeu1, which has recognition sites only in the 23S rRNA gene. The protocol for Southern hybridization method is provided in the website in the ‘About rrnDB’ section.
The new rrnDB also catalogues the number of internally transcribed spacer and tRNA genes per genome. The inclusion of the number of ITS helps capture organisms whose rRNA genes are not arranged in an operon, such as Leptospira sp., Thermoplasma sp. and Nanoarchaeum equitans (14–16). The rRNA genes of these organisms are separated on the chromosome and each under the control of their own promoter. The ITS region is increasingly used for diversity studies and since intragenomic heterogeneity increases with rrn operon copy number, the number of ITS region per genome will become more important in analyzing richness measures (17). Furthermore, organisms with multiple ITS can provide the heterogeneity required to differentiate organisms at subspecies level using restriction analysis (18),
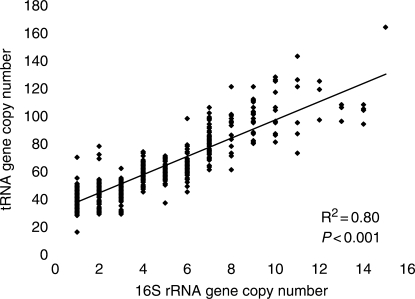
Amino-acylated tRNAs are substrates for ribosome-mediated protein synthesis. When selection favors an increased number of rrn genes to synthesize ribosomes more quickly, the rate of protein synthesis can only be increased if there is a corresponding increase in the production of tRNAs. A positive correlation between the abundance or tRNA and rRNA genes has been documented previously (19). A significant positive correlation is maintained in an analysis that is expanded to include 590 bacterial genomes (Figure 3). As expected, Photobacterium profundum, which has the highest number of 16S rRNA genes also has the highest number of total tRNA genes (20).
Figure 3.
Correlation between the total number of tRNA genes and 16S rRNA genes in bacterial genomes. The data are gathered from sequenced genomes of 590 bacterial species.
DATA CURATION
The rrnDB is equipped with a password protected entry form that is accessible from the WWW, allowing curators to update the database online at anytime. The authors currently handle curation and maintenance of the database. Genomic data are obtained from the NCBI Microbial Genomes database (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and the J. Craig Venter Institute Comprehensive Microbial Resource database (http://cmr.jcvi.org/tigr-scripts/CMR). Nongenomic data are obtained from literature searches, and these original references are maintained along with new genomic data and references as data from genome sequences are added. Taxonomic classifications are adopted from NCBI Entrez Taxonomy. The database is updated at least monthly.
The website has a ‘Contact Us’ form for users to alert the curators to new data for entry, ask questions about the site or provide suggestions for improving the rrnDB. The six most recent entries added to or updated in the database are listed in the left-side corner of the main page. Any changes in features are documented in the ‘news and updates’ section of the main page.
DATABASE MANAGEMENT SYSTEM
The rrnDB website is powered by PHP 5, MySQL 5, Apache 2 and runs on Mac OS X Server 10.4. The choice of programming language was made to facilitate ease of development and compatibility with available hardware. Except for the server operating system, all of these products are freely available under open source licenses, and have strong community support and a long history of integration.
The MySQL database is designed for speed and scalability: as new strains are added or updated through the administrative interface, the database size will grow, but by separating data out for as much normalization as possible, the growth will be reduced to a minimum. Database indexes and table structure make it fast to search by keyword or taxonomy classification.
The front-end website for users is designed for ease of use and utilizes AJAX technologies to make dynamic searching possible, including multi-level taxonomy, and seamlessly filters a list of matching strains when criteria are entered. Sorting of the result table is done by client-side JavaScript with no additional load on the server.
Another key feature of the front-end website is XHTML and CSS design, which makes it well-suited to technologies such as screen readers and other products that help make the site accessible to persons with disabilities. When such technologies do not have the capability to use JavaScript, the site automatically falls back to a version that will work in any browser, with no additional steps required by the user.
For site administrators, a graphical interface facilitates curation. Drop-down lists with journal names, taxonomy classifications, the ability to add multiple citations or chromosomes at once, and other fields make it simple and fast to enter or update data, and changes are immediately live on the site with no need to leave the browser. When data for new strains are entered or existing data are updated, timestamp fields in the database are updated, making it easy to search for new or changed information.
Overall, the site design emphasizes ease of use while still providing useful options for end users and efficient data entry and maintenance for administrators. It was our goal in designing this site that it should be usable by researchers for many years to come without needing to involve IT personnel for more than routine maintenance, and in the year it has been operational since its redesign, fewer than 10 hours have been spent by any IT personnel, either programmers or systems administrators, giving us confidence that this site is sustainable.
FUTURE PLANS
One planned addition to the rrnDB is information on intragenomic heterogeneity of rRNA genes. Although intragenomic gene conversion amongst copies of rrn genes maintains nearly identical sequences (21), differences between copies of rRNA genes are known. The highest intragenomic heterogeneity currently documented is 7.2% sequence divergence between 16S rRNA genes found in Thermobispora bispora (12). Documenting this variability will help estimate the contribution of intragenomic variation to the microheterogeneity that is frequently observed in environmental clone libraries of rRNA genes (22–24). The motivation to develop the rrnDB is to understand the evolutionary implication of redundancy of rrn genes, and so we also plan to expand the database to include genomic characteristics (e.g. gene content, pathway preferences) that correlate with the number of rRNA and tRNA genes.
FUNDING
National Science Foundation (IOS 0421900 to T.M.S.). Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank J.A. Klappenbach for the initial compilation of data.
REFERENCES
- 1.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 2.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crosby LD, Criddle CS. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques. 2003;34:790–794. doi: 10.2144/03344rr01. 796, 798 passim. [DOI] [PubMed] [Google Scholar]
- 6.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson BS, Schmidt TM. Growth rate-dependent accumulation of RNA from plasmid-borne rRNA operons in Escherichia coli. J. Bacteriol. 1998;180:1970–1972. doi: 10.1128/jb.180.7.1970-1972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichorst SA, Breznak JA, Schmidt TM. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 2007;73:2708–2717. doi: 10.1128/AEM.02140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavicchioli R, Ostrowski M, Fegatella F, Goodchild A, Guixa-Boixereu N. Life under nutrient limitation in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingomonas alaskensis) Microb. Ecol. 2003;45:203–217. doi: 10.1007/s00248-002-3008-6. [DOI] [PubMed] [Google Scholar]
- 10.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einen J, Thorseth IH, Ovreas L. Enumeration of archaea and bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy. FEMS Microbiol. Lett. 2008;282:182–187. doi: 10.1111/j.1574-6968.2008.01119.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Zhang Z, Ramanan N. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 1997;179:3270–3276. doi: 10.1128/jb.179.10.3270-3276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ojaimi C, Davidson BE, Saint Girons I, Old IG. Conservation of gene arrangement and an unusual organization of rRNA genes in the linear chromosomes of the Lyme disease spirochaetes Borrelia burgdorferi, B. garinii and B. afzelii. Microbiology. 1994;140(Pt 11):2931–2940. doi: 10.1099/13500872-140-11-2931. [DOI] [PubMed] [Google Scholar]
- 14.Fukunaga M, Masuzawa T, Okuzako N, Mifuchi I, Yanagihara Y. Linkage of ribosomal RNA genes in Leptospira. Microbiol. Immunol. 1990;34:565–573. doi: 10.1111/j.1348-0421.1990.tb01033.x. [DOI] [PubMed] [Google Scholar]
- 15.Ree HK, Zimmermann RA. Organization and expression of the 16S, 23S and 5S ribosomal RNA genes from the archaebacterium Thermoplasma acidophilum. Nucleic Acids Res. 1990;18:4471–4478. doi: 10.1093/nar/18.15.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, Beeson KY, Bibbs L, Bolanos R, Keller M, et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl Acad. Sci. USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart FJ, Cavanaugh CM. Intragenomic variation and evolution of the internal transcribed spacer of the rRNA operon in bacteria. J. Mol. Evol. 2007;65:44–67. doi: 10.1007/s00239-006-0235-3. [DOI] [PubMed] [Google Scholar]
- 18.Malimas T, Yukphan P, Takahashi M, Potacharoen W, Tanasupawat S, Nakagawa Y, Tanticharoen M, Yamada Y. Heterogeneity of strains assigned to Gluconobacter frateurii Mason and Claus 1989 based on restriction analysis of 16S-23S rDNA internal transcribed spacer regions. Biosci. Biotechnol. Biochem. 2006;70:684–690. doi: 10.1271/bbb.70.684. [DOI] [PubMed] [Google Scholar]
- 19.Dethlefsen L, Schmidt TM. Performance of the translational apparatus varies with the ecological strategies of bacteria. J. Bacteriol. 2007;189:3237–3245. doi: 10.1128/JB.01686-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vezzi A, Campanaro S, D’Angelo M, Simonato F, Vitulo N, Lauro FM, Cestaro A, Malacrida G, Simionati B, Cannata N, et al. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science. 2005;307:1459–1461. doi: 10.1126/science.1103341. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto JG, Stevenson BS, Schmidt TM. Rates and consequences of recombination between rRNA operons. J. Bacteriol. 2003;185:966–972. doi: 10.1128/JB.185.3.966-972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JBH. It's all relative: ranking the diversity of aquatic bacterial communities. Environ. Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- 23.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]