Abstract
Volatiles are efficient mediators of chemical communication acting universally as attractant, repellent or warning signal in all kingdoms of life. Beside this broad impact volatiles have in nature, scents are also widely used in pharmaceutical, food and cosmetic industries, so the identification of new scents is of great industrial interest. Despite this importance as well as the vast number and diversity of volatile compounds, there is currently no comprehensive public database providing information on structure and chemical classification of volatiles. Therefore, the database SuperScent was established to supply users with detailed information on the variety of odor components. The version of the database presented here comprises the 2D/3D structures of approximately 2100 volatiles and around 9200 synonyms as well as physicochemical properties, commercial availability and references. The volatiles are classified according to their origin, functionality and odorant groups. The information was extracted from the literature and web resources. SuperScent offers several search options, e.g. name, Pubchem ID number, species, functional groups, or molecular weight. SuperScent is available online at: http://bioinformatics.charite.de/superscent.
INTRODUCTION
In scientific terms, scents are mixtures of volatile compounds with a high vapor pressure and a molecular weight, which is usually <300 g mol−1 (1). Human beings often associate scents with volatiles that can be perceived by the human nose and have a pleasant smell. But the entire group of volatile compounds comprises thousands of inorganic and organic compounds stemming from major pathways of secondary metabolisms of many organisms (1). These volatiles may affect living organisms in one way or the other. Over the past decades, scientific investigations have revealed that volatiles play a key role in life by acting as semiochemicals, mediating inter- and intraspecies interactions of living organisms.
Volatiles allow animals to recognize or detect individuals. The volatiles called pheromones are indispensable for mating choices, sexual behavior and fertilization, and also for nursing. They are important for the maintenance of social relationships, especially in animal communities such as hives, ant colonies, prairie dog towns or even packs/prides/herds of larger animals. Volatiles also support foraging and the detection of prey. They can also serve as signals to warn kin in situations of danger (alarm pheromones) or they are even used to defend against predators. The relevance of volatiles is not restricted to the animal kingdom. Volatile semiochemicals are involved in plant–plant interactions (2) and many play a crucial role in plant–animal interactions, e.g. pollination (3), herbivory (4) and the plants’ response to defend against herbivores (5). Recently, it was shown that bacteria also emit a wealth of volatiles with an impact on plants, fungi, animals and bacteria (M. Kai et al., Bacterial volatiles and their action potential, Applied Microbiology Biotechnology, submitted for publication).
Last but not least, scent components are of tremendous commercial interest, resulting in many applications of volatiles in science and industry. Certain pleasant odors have a positive effect on customer and are therefore used in shopping malls, for wellness applications, and for the production of perfumes, cosmetics and household cleaning agents, to name a few.
For the recognition of odorant molecules, a large variety of olfactory receptors is known in humans and animals. To discriminate between scent components, each receptor has affinities for a range of molecules and, by combination of activated receptors, many different smells can be perceived (6). All olfactory receptors are members of class A rhodopsin-like family of G-protein-coupled receptors (GPCRs) (7), a group of seven transmembrane domain receptors activating signal transductions in cells. Crystal structures are currently available for rhodopsin (8) and β2-adrenoreceptor (9). An excellent source for information about olfactory receptors is the Olfactory Receptor database (10). This database provides information about the perceiving proteins and complexes, and a database gathering comprehensive knowledge on volatiles should give information about their ligands. The work of Schmuker et al. (11,12) bridges the gap between the structure of volatile compounds and the receptor response by prediction via machine learning techniques using experimental data from in vivo receptor recordings.
Hitherto, a comprehensive compiliation of volatiles was not publicly available for scientific use. It was therefore our goal to establish the database ‘SuperScent’. A variety of database resources of scent components is already known, but they are limited in scope and focus on certain subgroups of scents, e.g. the OdorDB, part of the neuroscience Senselab project (13), centers on odorant molecules experimentally shown to bind olfactory receptor proteins, whereas the Pherobase database (http://www.pherobase.com/) focuses on pheromones but meanwhile also covers a broad variety of semiochemicals. A compilation of floral scent components is found in the ScentBase (http://www2.dpes.gu.se/SCENTbase.html) and the Flavornet (http://www.flavornet.org/flavornet.html) summarizes volatile compounds found in the human olfactory perception space. Consequently, these databases are useful for special purposes, but there is still a need for a comprehensive listing of volatiles regarding their properties and their uses in science and industry.
The SuperScent database, which is presented here, comprises more than 2100 compounds, together with several classification criteria. These volatile compounds were collected from a variety of sources, e.g. the literature or other databases, leading to a comprehensive dataset of scent components, together with information about their chemical properties and commercial availability. These features provide the user with an extensive database, together with substantial options, such as a search within particular group odor subgroups.
THE DATABASE
With 2147 compounds, 9214 synonyms and references to more than 20 different suppliers, SuperScent provides the largest diversity of volatile compounds and corresponding information available online. The data are accessible with two different search options: ‘Scent Search’ and ‘Structure Search’.
The ‘Scent Search’ enables the user to look for compounds by means of the PubChem-ID or the name. Additionally, by choosing certain functional groups, species, or range of molecular weights, all database entries meeting the search criteria can be accessed.
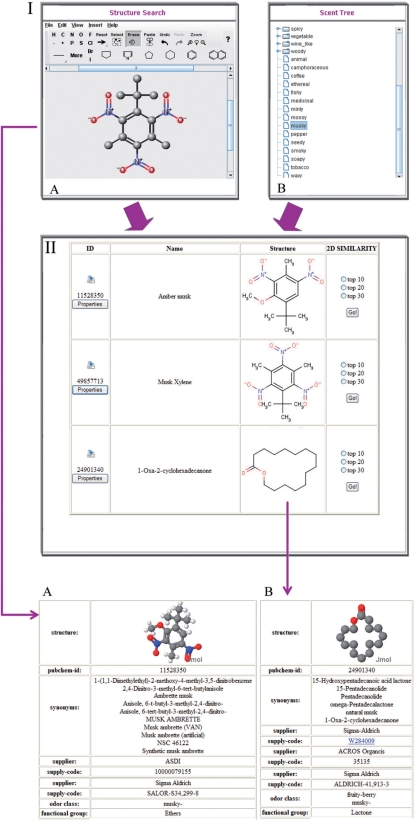
The second way to screen the database requires a molecular structure (‘Structure Search’). Here, a SMILES code (Simplified Molecular Input Line Entry System) or a MOL-file of the search compound can be uploaded. With the help of MarvinView (http://www.chemaxon.com), the user can draw either the whole structure of the search molecule, or parts of it (I-A in Figure 1). Thus, it is possible to screen the database with self-edited molecule structures. In order to find resembling structures in the database, a similarity search is performed. The 10 most similar database entries are listed in the order of similarity (II in Figure 1). For each compound, the name, the PubChem-ID, the 2D structure and the Tanimoto coefficient are presented. Furthermore, a similarity search to find the 10, 20 or 30 most similar compounds is provided.
Figure 1.
Flow chart of a scent search in the SuperScent database. (I) Different search options are provided: (A) Structure search: upload an MOL-file or SMILES code, but it is also possible to draw one's own structure. (B) Scent tree: structures are clustered into scent classes. The branch can be expanded by clicking on the nodes and a click on one subclass shows a list of compounds. (II) Result table of a scent search. The detailed view for two molecules is depicted: (A) synthetic musk. (B) natural musk.
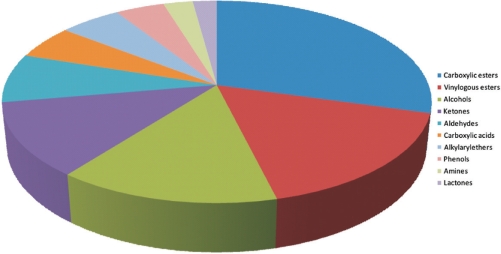
Additional information is available in a separate ‘Properties’ window (II-A/B in Figure 1). The user can find synonyms, functional groups, the molecular weight and all species in which the compound has been found, together with the corresponding reference. Furthermore, the compounds are classified according to their structure (e.g. benzenoids), chemical features (Figure 2), quality of scent (e.g. fruity-peach) and ordering information (supplier; ID).
Figure 2.
Pie chart of chemical classes found in the SuperScent database.
Another useful feature of the web interface is the ‘Scent Tree’ (I-B in Figure 1). Here, the database entries have been clustered according to the quality of their aroma. There are 29 classes, such as balsamic, floral or spicy, which are again divided into several subclasses. For instance, the fruity scents include subclasses such as apple, banana or coconut, leading to an overall number of 121 different groups.
A manually verified upload option allows the scientific community to contribute to the database. Here, the user can import a MOL-file together with corresponding information of the compound. The SuperScent database will be updated twice a year.
METHODS
Data were collected from the literature and various web resources, such as a collection of floral scents (1), a review outlining bacterial volatiles (14) and PubChem (http://pubchem.ncbi.nlm.nih.gov/). Abstracts of the literature database PubMed were filtered for relevant articles using specific keywords. The abstracts were screened against names and synonyms of chemical compounds, as well as a distinct set of substrings of IUPAC names. The text passages containing matches were manually curated by a scientific team of biologists that confirmed the matching compounds and verified them. Several web resources, e.g. the Riechstoff-Lexikon (http://omikron-online.de) and the Flavors and Fragrances catalog (http://safcsupplysolutions.com) supplied by Sigma Aldrich were checked.
SuperScent is designed as a relational database, which is implemented on a MySQL server. For chemical functionality, the MyChem package, which aims to provide a complete set of functions for handling chemical data within MySQL, is added. Most of the functions used by MyChem depend upon Open Babel (15). The fingerprint algorithm implemented in Open Babel follows the Daylight approach (http://www.daylight.com/dayhtml/doc/theory/). As similarity index, the Tanimoto coefficient is used, calculating the number of bit positions set to 1 in both fingerprints, divided by the number of bit positions set to 1, in at least one of the fingerprints. If a set bit is considered as a feature present in the molecule, the Tanimoto coefficient is a measure of the number of common features in both molecules (16). A Tanimoto coefficient of >0.85 indicates that two molecules may have similar activities (17). For displaying 3D structures, Jmol, an open-source Java viewer for chemical structures in 3D (http://www.jmol.org/) is used. Marvin ChemSketch was applied for the built-in molecule editor, which allows structural screening with self-edited molecules. The website is built with php and javascript, and web access is enabled via Apache HTTP Server 2.2.
CONCLUSION AND FUTURE DIRECTION
The SuperScent database has become a useful tool to retrieve information about scents or to get an overview of the known volatile organic compounds. The included data on purchasability of scents will enable systematic experimental approaches on the relation between structural similarities and scent classes. Furthermore, structure comparisons of self-edited molecules with the annotated scents may allow a first rough estimation of the potential aroma of new chemicals. The SuperScent database is a free resource with embedded screening functions for chemical compounds. The extension of the database allows the scientific community simple access to a growing number of available scents. We plan to extend the database by including olfactory receptors in the near future.
FUNDING
Deutsche Forschungsgemeinschaft (SFB 449); Investitionsbank Berlin (IBB); International Research Training Group (IRTG); Berlin-Boston-Kyoto and Deutsche Krebshilfe. Funding for open access charge: SFB 449.
Conflict of interest statement. None declared.
REFERENCES
- 1.Knudsen JT, Eriksson R, Gershenzon J, Ståhl B. Diversity and distribution of floral scent. Bot. Rev. 2006;72:1–120. [Google Scholar]
- 2.Baldwin IT, Halitschke R, Paschold A, von Dahl CC, Preston CA. Volatile signaling in plant-plant interactions: “talking trees” in the genomics era. Science. 2006;311:812–815. doi: 10.1126/science.1118446. [DOI] [PubMed] [Google Scholar]
- 3.Dobson HEM. Relationship between floral fragrance composition and type of pollinator. In: Dudareva N, Pichersky E, editors. Biology of Floral Scent. Boca Raton, FL: Taylor and Francis; 2006. pp. 147–198. [Google Scholar]
- 4.Vet LEM, Dicke M. Ecology of infochemical use by natural enemies in a tritrophic context. Annu. Rev. Entomol. 1992;37:141–172. [Google Scholar]
- 5.Heil M, Silva Bueno JC. Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc. Natl Acad. Sci. USA. 2007;104:5467–5472. doi: 10.1073/pnas.0610266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- 7.Krautwurst D. Human olfactory receptor families and their odorants. Chem. Biodivers. 2008;5:842–852. doi: 10.1002/cbdv.200890099. [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 9.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crasto C, Marenco L, Miller P, Shepherd G. Olfactory Receptor Database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res. 2002;30:354–360. doi: 10.1093/nar/30.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmuker M, de Bruyne M, Hahnel M, Schneider G. Predicting olfactory receptor neuron responses from odorant structure. Chem. Cent. J. 2007;1:11. doi: 10.1186/1752-153X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmuker M, Schneider G. Processing and classification of chemical data inspired by insect olfaction. Proc. Natl Acad. Sci. USA. 2007;104:20285–20289. doi: 10.1073/pnas.0705683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crasto CJ, Marenco LN, Liu N, Morse TM, Cheung KH, Lai PC, Bahl G, Masiar P, Lam HY, Lim E, et al. SenseLab: new developments in disseminating neuroscience information. Brief. Bioinform. 2007;8:150–162. doi: 10.1093/bib/bbm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 2007;24:814–842. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- 15.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The Blue Obelisk-interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaney JS. Assessing the ability of chemical similarity measures to discriminate between active and inactive compounds. Mol. Divers. 1996;1:217–222. doi: 10.1007/BF01715525. [DOI] [PubMed] [Google Scholar]
- 17.Patterson DE, Cramer RD, Ferguson AM, Clark RD, Weinberger LE. Neighborhood behavior: a useful concept for validation of “molecular diversity” descriptors. J. Med. Chem. 1996;39:3049–3059. doi: 10.1021/jm960290n. [DOI] [PubMed] [Google Scholar]