Abstract
Ligands that activate the epidermal growth factor receptor (EGFR) are synthesized as membrane-anchored precursors that appear to be proteolytically released by members of the ADAM family of metalloproteases. Because membrane-anchored EGFR ligands are thought to be biologically active, the role of ligand release in the regulation of EGFR signaling is unclear. To investigate this question, we used metalloprotease inhibitors to block EGFR ligand release from human mammary epithelial cells. These cells express both transforming growth factor α and amphiregulin and require autocrine signaling through the EGFR for proliferation and migration. We found that metalloprotease inhibitors reduced cell proliferation in direct proportion to their effect on transforming growth factor α release. Metalloprotease inhibitors also reduced growth of EGF-responsive tumorigenic cell lines and were synergistic with the inhibitory effects of antagonistic EGFR antibodies. Blocking release of EGFR ligands also strongly inhibited autocrine activation of the EGFR and reduced both the rate and persistence of cell migration. The effects of metalloprotease inhibitors could be reversed by either adding exogenous EGF or by expressing an artificial gene for EGF that lacked a membrane-anchoring domain. Our results indicate that soluble rather than membrane-anchored forms of the ligands mediate most of the biological effects of EGFR ligands. Metalloprotease inhibitors have shown promise in preventing spread of metastatic disease. Many of their antimetastatic effects could be the result of their ability to inhibit autocrine signaling through the EGFR.
The epidermal growth factor receptor (EGFR) plays an important role during development. Knockout of the EGFR gene results in numerous developmental abnormalities in the brain, skin, and gut (1, 2). A variety of ligands in addition to EGF have been shown to stimulate the EGFR, including transforming growth factor α (TGFα) (3), amphiregulin (AR) (4), heparin-binding EGF (5), and betacellulin (6). All of these ligands are made as membrane-spanning prohormones that are processed and released through regulated proteolysis (7). Although the identities of all of the proteases involved have not been definitively established, recent data suggests that the release of TGFα involves TACE, a member of the ADAM family of metalloproteases, which originally was identified as being responsible for the release of tumor necrosis factor α (8–10).
Disruption of the EGFR gene in mice indicates that epithelial cells are most profoundly affected by receptor loss (1, 2, 11). Interestingly, knockout of the TACE gene in mice results in a very similar phenotype (10). Although these data have been interpreted to indicate that proteolytic release of EGFR ligands is important in receptor function in vivo, this conclusion is contradicted by numerous in vitro studies that appear to show that membrane-anchored growth factors are biologically active in a juxtacrine fashion (12–14). One possible explanation is that the activities of membrane-anchored ligands are distinct from those of the soluble forms. Perhaps juxtacrine ligands mediate short-range signaling, whereas soluble ligands operate on distal cells. Alternatively, the in vitro studies on membrane-anchored ligands may have been misleading. Those studies typically have used artificial systems in which the cell expressing the ligand is distinct from the cell expressing the receptor (12–14). Most EGF-dependent cells, such as those found in the gut, kidneys, and epidermis, have been shown to express one or more EGFR ligands in an autocrine fashion (15–17). In addition, EGFR ligands stimulate a number of different biological responses in these cells, such as proliferation and migration, which may display different sensitivities to membrane-anchored versus soluble ligand (18). Because of limitations in previous experimental systems, the relative activities of soluble versus membrane-anchored ligands have been difficult to compare.
The release of several EGFR ligands can be blocked by low molecular weight hydroxamate compounds, which are selective metalloprotease inhibitors (19–21). One of these inhibitors, batimastat, has been used in clinical trials as an inhibitor of tumor metastasis (22, 23). Although batimastat initially was thought to work by inhibiting matrix-degrading enzymes, such as collagenase, recent studies indicate that its mode of action is more complex (24). To determine whether batimastat exerts some of its antimetastatic effects by inhibiting release of EGFR ligands, we used a human mammary epithelial cell line (HMEC line 184A1) that previously has been shown to depend on autocrine signaling through the EGFR for growth and proliferation (25). Here, we show that blocking the proteolytic release of EGFR ligands essentially abolishes their biological activities, suggesting that at least some membrane-anchored forms of EGFR ligands are functionally inactive. In addition, the efficiency at which batimastat blocks proliferation and migration of epithelial cells suggests that much of its antimetastatic activity could be mediated by interference with autocrine signaling through the EGFR system.
MATERIALS AND METHODS
General.
HMEC line 184A1 (26) was obtained from Martha Stampfer (Berkeley National Laboratory) and cultured in medium DFCI-1 as described (27). HCT-116 cells were obtained from the American Type Culture Collection. Cells expressing EGF with carboxyl terminus (EGF-Ct) and secreted EGF (sEGF) have been described (18). mAb 225 directed against the EGFR (28) was isolated from a hybridoma cell line obtained from the American Type Culture Collection. EGF was obtained from Peprotech (Rocky Hill, NJ). Anti-phosphotyrosine horseradish peroxidase conjugate (RC-20) and anti-EGFR antibody (C-13) used in Western blots were obtained from Transduction Laboratories, Lexington, KY. Batimastat (BB-94) and BB-2116 were obtained from British Biotechnology (Oxford, U.K.). Matrix metalloproteinase 3 inhibitor was obtained from Calbiochem.
Ligand Cleavage.
Cells were plated into 60-mm dishes and grown until near confluence. The medium was replaced with 2 ml of fresh medium containing the appropriate inhibitor together with 20 μg/ml of 225 mAb to block ligand uptake by the cells. After 18 hr, the medium was harvested for determination of ligand concentration. Cells were counted, and ligand concentration was normalized to nanograms per million cells. EGF concentration was determined by ELISA as described (29). Concentrations of TGFα were determined by RIA as described (30). Concentrations of AR were determined by sandwich ELISA. Capture antibody (anti-AR 6RIC 2.4) was absorbed to wells overnight (0.2 μg/well) and blocked with 3% BSA and 0.5% Tween-20. Samples were incubated for 1 hr at 37°C and secondary antibody (0.1 μg/well biotinylated anti-AR 4.14.18) was added for 1 hr at 37°C followed by 0.05 μg/ml of peroxidase-conjugated streptavidin. Substrate (0.5 mg/ml of O-phenylenediamine dihydrochloride) was added for 5 min, and the reaction product was read in a microplate reader. Standards were recombinant human AR (R&D Systems) diluted in culture medium.
EGFR Phosphorylation.
Cells were plated into 100-mm dishes and grown until near confluence and then treated with or without 10 μg/ml of mAb 225 or 10 μM batimastat for 24 hr. EGF (100 ng/ml) was added to one set of plates for 20 min. Cell extracts were isolated in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) in the presence of 1 mM orthovanadate and protease inhibitors (31). Equal amounts of protein were immunoprecipitated with mAb 225, rabbit anti-mouse IgG, and protein A-Sepharose. The immunoprecipitates were resolved on 5–10% SDS/PAGE gels and transferred to nitrocellulose membrane. The membrane was probed with horseradish peroxidase-conjugated antiphosphotyrosine antibody (RC-20), followed by ECL detection (Amersham Pharmacia). Band density was determined by using a Bio-Rad model GS-670 Imaging Densitometer. After stripping with Tris buffer containing 2% SDS, the blots were reprobed with anti-EGFR mAb C-13 and goat anti-mouse IgG horseradish peroxidase conjugate.
Proliferation Assays.
Cells were plated on coverslips and grown in complete DFCI-1 medium for 1 day. Then the cells were switched to control medium, or medium containing the indicated additives for 2 days. BrdUrd (10 μM) was added for the last 18 hr to label any cells in S phase. Cells were stained with a BrdUrd labeling and detection kit (Boehringer Mannheim) and counterstained with 15 nM 4′,6-diamidino-2-phenylindole (DAPI). Random fields of cells were selected by using an automated stage operated with openlab software (Improvision, Boston). Images of nuclei were obtained at 470 nm (DAPI) and 520 nm (BrdUrd) and were counted by using the openlab density slicing and automator modules. At least 20 random fields were counted for each slide. Selected manual counts were done to confirm the accuracy of the software. For direct determination of cell proliferation, cells were plated at 5,000–10,000 per well in 24-well dishes. The next day the cells were changed to growth medium with the indicated additions. Two days later, the medium was changed, and after an additional 2 days the cells were removed from the dishes by trypsin, and cell numbers were determined by using a Coulter counter.
Clonal Density Growth.
Cells were plated into 35-mm dishes at clonal density (1:400) and grown in complete DFCI-1 medium or DFCI-1 medium supplemented with 10 μM batimastat. The medium was changed every 2 days. After 2 weeks, plates were stained with Giemsa.
Cell Tracking.
Confluent cultures of cells split at a 1:50 ratio 20 hr previously were treated either with or without either EGF or batimastat for 4 hr and mounted on a 37°C heated stage by using DFCI-1 medium lacking bicarbonate but containing 25 mM Hepes. The medium was overlaid with mineral oil to prevent evaporation. Phase contrast images were taken of random fields at 10-min intervals for a total of 15 hr by using a 10× objective and openlab software (Improvision). Cells were marked manually in the center of the nucleus at each time point by using the Advanced Measurements module. The mean squared displacement of the cells as a function of time was calculated as described (32).
RESULTS
Batimastat Inhibits Proliferation of EGF-Dependent Cells.
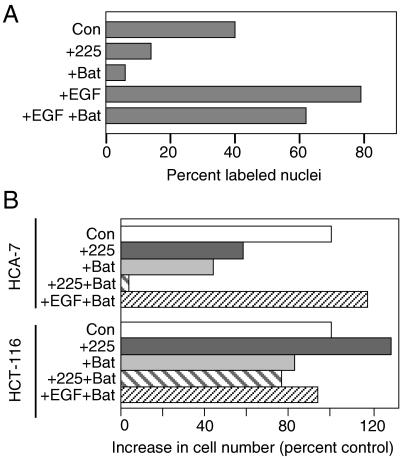
To determine whether metalloprotease inhibitors could reduce proliferation of HMEC, we treated cells either with or without batimastat for 48 hr. During the final 18 hr, BrdUrd was added to label cells in S phase. As shown in Fig. 1A, the addition of batimastat reduced the percentage of BrdUrd-labeled cells from 40% to 6%. Similarly, blocking endogenous autocrine EGFR signaling with the antagonistic anti-EGFR mAb 225 reduced the percentage of labeled cells to 14%. The addition of soluble EGF stimulated proliferation of the cells in either the presence or absence of batimastat. Thus, although batimastat inhibited proliferation of HMEC, soluble EGF appeared to reverse most of this effect.
Figure 1.
Batimastat (Bat) inhibits proliferation of EGF-dependent cells. (A) HMEC strain 184A1 were plated on coverslips and treated with mAb 225 (67 nM), batimastat (10 μM), or EGF (50 ng/ml) for 48 hr. Cells were labeled during the last 18 hr with BrdUrd. The results are the average of two independent experiments. (B) The indicated cells were grown for 5 days in growth medium alone (control), the presence of mAb 225 (67 nM), batimastat (7.5 μM), or a combination of mAb 225 and batimastat or batimastat and EGF (4 nM). Media were changed on days 1 and 3.
To determine whether batimastat inhibits the proliferation of other cells that depend on autocrine signaling through the EGFR, we compared its effect on the colon cancer lines HCA-7 and HCT-116. The growth of HCA-7 cells previously has been shown to be inhibited by blocking the EGFR whereas HCT-116 cells are unresponsive to EGF (33, 34). As shown in Fig. 1B, the addition of either batimastat or mAb 225 alone inhibited the growth of HCA-7 cells by about 60%. The addition of both mAb 225 and batimastat completely blocked the growth of these cells. Very similar results were obtained by using the autocrine-dependent MDA-MB-468 cell line (data not shown). The addition of EGF reversed the effect of batimastat on HCA-7 cells. The growth of HCT-116 cells, however, was not inhibited by the addition of either mAb 225 or batimastat or a combination of the two reagents. These results indicate that cells that are inhibited by blocking the EGFR also are inhibited by batimastat.
We evaluated the levels of mRNA encoding the different EGFR ligands in HMEC by using the reverse transcriptase–PCR technique and found significant levels of only TGFα and AR (data not shown). We then verified that metalloprotease inhibitors can inhibit the release of these ligands from HMEC. We found that during an 18-hr treatment period, 5 μM batimastat reduced the amount of TGFα found in the medium from 86 pg/ml to undetectable levels whereas AR was reduced from 3.5 to 1.6 ng/ml. Thus it appears that batimastat treatment can inhibit release of both TGFα and AR.
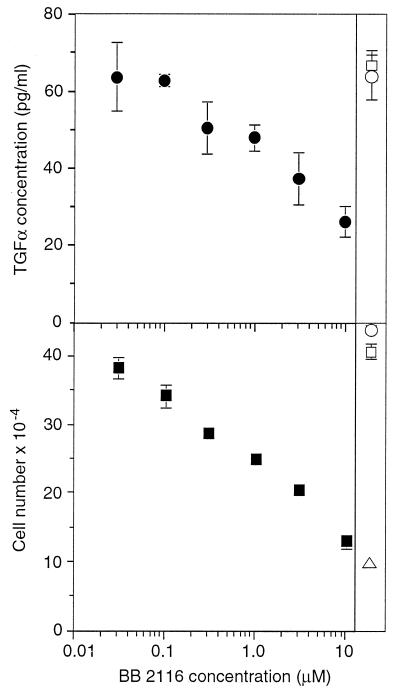
To explore the relative contribution of TGFα versus AR release in HMEC proliferation, we evaluated several different metalloprotease inhibitors for their ability to differentially inhibit release of TGFα versus AR. We found that the compound BB-2116, a water-soluble derivative of batimastat, appeared to be a relatively selective inhibitor of TGFα release. At a concentration of 10 μM, BB-2116 reduced the amount of TGFα found in the medium during an 18-hr period from 64 to 26 pg/ml whereas AR was unaltered (from 0.53 to 0.52 ng/ml in the presence and absence of 10 μM BB-2116, respectively). We then treated cells with varying concentrations of BB-2116 and measured both TGFα release and cell proliferation. As shown in Fig. 2, increasing concentrations of BB-2116 caused a dose-dependent reduction in the amount of TGFα released from HMEC. Cell proliferation also was reduced in direct proportion to the reduction in TGFα release. Metalloprotease inhibitors that had no effect on TGFα release (such as matrix metalloproteinase 3 inhibitor) had no effect on cell proliferation. Therefore, the effects of metalloprotease inhibitors on HMEC proliferation appears to be directly correlated to their effects on TGFα release.
Figure 2.
Inhibition of cell proliferation is correlated with inhibition of TGFα release. (Upper) HMEC strain 184A1 were grown to near confluency and changed to medium containing the indicated concentration of BB-2116 and 20 μg/ml of mAb 225 to prevent ligand uptake by the cells. After 18 hr, the medium was collected and evaluated for TGFα concentration, normalized to 106 cells and ± SD. As a control, cells were also incubated with 50 μM matrix metalloproteinase 3 inhibitor (□) or with no inhibitor (○). (Lower) Cells were split 1:10 into 12-well dishes, and 18 hr later were changed to medium containing the indicated concentration of BB-2116. The medium was changed every 2 days and cells were counted on day 6. Shown are the results of duplicate wells ± SD. Controls are same as Upper as well as 20 μg/ml mAb 225 (▵).
Cells Expressing Soluble EGF Are Not Inhibited by Batimastat.
The EGFR system is “autoinductive” in that EGFR activation can stimulate the synthesis of EGFR ligands (16). It seemed possible that the effect of batimastat on cell proliferation might not be caused directly by inhibition of EGFR ligand release, but instead caused by an effect on EGFR signaling and a consequent reduction in ligand gene expression. Direct measurements did not support this hypothesis, however. The amount of TGFα found in cell lysates was not reduced after batimastat treatment (from 950 pg to 1,200 pg per 106 cells before and after 5 μM batimastat treatment for 18 hr, respectively).
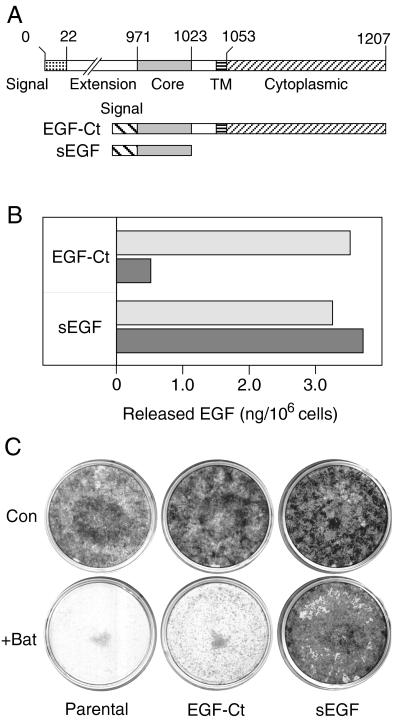
If batimastat is preventing cell proliferation by inhibiting release of membrane-anchored EGFR ligands, then we should be able to circumvent this inhibition by removal of the membrane anchor. We therefore engineered an artificial EGF gene that lacks transmembrane and cytoplasmic domains (Fig. 3A; also see ref. 29). Unlike native EGF, the product of this artificial gene (sEGF) does not require proteolytic processing. To make a control construct, sequences of the transmembrane and cytoplasmic domains of EGF were added to yield an artificial membrane-anchored form of EGF (EGF-Ct). The product of this second artificial gene still should require proteolysis to be released from the cell surface. These two EGFR ligands then were stably expressed in HMEC using retrovirus-mediated gene transfer (35). Because these constructs use retroviral promoters, their expression levels are independent of EGFR activity.
Figure 3.
Batimastat inhibits both release and mitogenic activity of a membrane-anchored, but not a soluble form of EGF. (A) Map of the artificial EGF genes expressed in HMEC. (Upper) The native EGF gene from which the two artificial genes were derived. (B) Cells expressing either the EGF-Ct or sEGF constructs were incubated with 67 nM mAb 225 (to prevent ligand uptake) for 18 hr either without (empty bar) or with (filled bar) 5 μM batimastat. The results are the average of two independent experiments. (C) Parental cells and those expressing the indicated construct were plated at a 1:400 dilution and grown for 2 weeks either with or without 10 μM batimastat (Bat). The medium was changed every 2 days. Cells then were stained with Giemsa.
In the absence of batimastat, cells expressing either sEGF or EGF-Ct released similar amounts of EGF into the medium (Fig. 3B). Batimastat reduced the amount of EGF released from cells expressing EGF-Ct by 86% during an 18-hr period. In contrast, there was no effect on the release of EGF from cells expressing sEGF. Additional experiments demonstrated that the concentration of batimastat necessary for half-maximum inhibition of EGF release from the cells expressing EGF-Ct was approximately 0.5–1 μM. We conclude that batimastat effectively inhibits the release of membrane-anchored EGF, most likely through its effect on the processing protease. This inhibition is very similar to what has been described for the native EGF molecule (21).
To determine whether batimastat can selectively block the growth and proliferation of cells expressing membrane-anchored EGF, we seeded HMEC expressing either sEGF or EGF-Ct, as well as the parental cell line, at clonal density in either the presence or absence of batimastat. After 14 days in culture, the plates were stained to visualize cell colonies. As shown in Fig. 3C, batimastat effectively blocked the clonal growth of both the parental HMEC as well as cells expressing EGF-Ct. Significantly, batimastat had little effect on cells expressing secreted sEGF. Results from BrdUrd labeling studies confirmed these results (data not shown). The addition of batimastat reduced the percent of cells entering S phase from 78% to 25% in the case of cells expressing EGF-Ct, but had no effect on cells expressing sEGF (80% and 78% in the presence and absence of batimastat, respectively). The addition of exogenous EGF reversed the effect of batimastat on cells expressing EGF-Ct (from 25% to 74%; data not shown). Although batimastat (or soluble EGF) had no effect on the behavior of cells expressing sEGF, these cells still depended on EGFR activation. The addition of PD 153035, a specific inhibitor of the EGFR kinase (36), effectively blocked the growth of these cells (data not shown). Together, these results show that batimastat selectively inhibits the proliferation of cells expressing membrane-anchored forms of EGFR ligands.
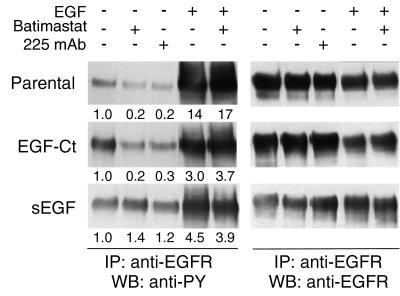
If batimastat is preventing HMEC proliferation through its effect on EGF release, we also should observe an inhibition of EGFR activation. To directly evaluate this prediction, cells were treated with batimastat, antagonistic mAb 225 or EGF, and the level of EGFR tyrosine phosphorylation was determined by Western blots. As shown in Fig. 4, the presence of batimastat inhibited autocrine activation of the EGFR in both parental HMEC and cells expressing EGF-Ct, but had no effect on cells expressing sEGF. The degree of inhibition with batimastat was similar to that observed for the antagonistic mAb 225. The activation of EGFR by sEGF could not be efficiently inhibited by mAb 225 because sEGF binds to the EGFR before receptor arrival at the cell surface (18). Addition of a bolus of soluble EGF resulted in a high level of EGFR phosphorylation that was not affected by the presence of batimastat. These results show that batimastat has no direct effect on EGFR activation, but appears to work by inhibiting ligand release.
Figure 4.
Batimastat blocks autocrine signaling of cleavage-dependent EGFR ligands. Confluent cultures of either parental HMEC or cells expressing EGF-Ct or sEGF were preincubated for 24 hr with either 67 nM mAb 225 or 10 μM batimastat. Treatment with EGF (100 ng/ml) was for 20 min. Total EGFR was immunoprecipitated (IP) and visualized by Western blot (WB) using anti-phosphotyrosine (anti-PY) antibodies. The blots were then stripped and reprobed with anti-EGFR antibodies. The numbers under the lanes are the relative densities of the bands normalized to the untreated controls.
Batimastat Inhibits Migration of Epithelial Cells.
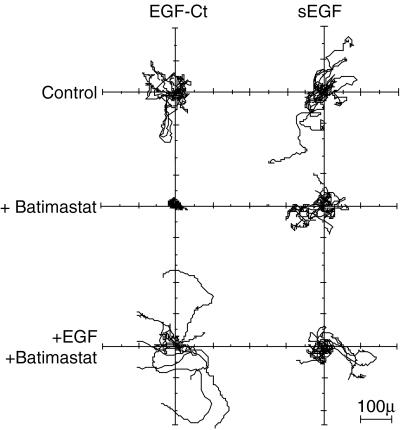
Batimastat previously has been shown to inhibit the ability of cells to invade basement membranes, presumably because of its ability to inhibit matrix-degrading enzymes (24, 37). Cell invasiveness, however, also requires active migration of the cells into a matrix. Because EGF is known to stimulate the motility a variety of epithelial cells (38, 39), we hypothesized that batimastat also might inhibit cell migration by blocking EGFR ligand release. We tested this hypothesis by investigating the movement of cells in either the presence or absence of batimastat. Cell movement was followed by time-lapse video microscopy for 15 hr in the presence or absence of batimastat. The position of the cells then was recorded and normalized to their starting position. The tracking data for eight cells randomly chosen from each group is shown in Fig. 5. It is clear that batimastat strongly inhibited the migration of cells expressing EGF-Ct, but had little effect on cells expressing sEGF. Batimastat also inhibited the migration of parental HMEC (data not shown). As was the case with proliferation, the addition of exogenous EGF reversed the inhibitory effect of batimastat (Fig. 5).
Figure 5.
Inhibition of cell migration by batimastat. HMEC expressing the indicated ligand were followed for 15 hr by time-lapse video microscopy using 10-min intervals. The tracks of eight random cells from each plate are plotted as flower plots (32) with the origin of each cell set to 0,0. Batimastat was used at 10 μM, and EGF was used at 50 ng/ml.
Batimastat reduced the speed of cells expressing EGF-Ct and sEGF by 50% and 18%, respectively (from 38 to 19 μm × h−1 and from 34 to 28 μm × h−1). Reliable values for directional persistence could not be determined in the presence of batimastat. This inhibitor caused the cell migration behavior to become directionally erratic, yielding tracking data that could not be fit to the persistent random walk model used for the analysis (32). Visual observations showed that cells treated with batimastat tended to “oscillate” around their initial location with little net translocation, indicating that batimastat has a strong negative effect on cell persistence. This idea is supported by inspection of the tracking data (compare the top left and middle left panels of Fig. 5). The net effect of reducing both speed and persistence would be an inability of cells to migrate any significant distance from their origin. Because exogenous or “preprocessed” EGF readily reverses the effect of batimastat (Fig. 5), it appears that batimastat primarily inhibits cell migration by interfering with EGFR ligand release.
DISCUSSION
Ever since the discovery that EGFR ligands are made as membrane-anchored precursors, the functional role of this form has been debated. The current paradigm is that membrane-anchored growth factors act as juxtacrine regulators of cell-cell signaling (7). This conclusion was based on the demonstration that cells expressing protease-resistant forms of ligands such as TGFα, could activate receptors on neighboring cells (12, 13). Although these studies demonstrated clear evidence of cell-cell signaling, several caveats remain with respect to their interpretation. First, cleavage resistance of the constructs was evaluated by the lack of detectable TGFα in the medium (13). Subsequently, however, it has been shown that released EGFR ligands are not necessarily detectable because of the high capture efficiency of EGFR on the ligand-expressing cells (18, 30). We have, in fact, found significant amounts of TGFα released by cells expressing “noncleavable” TGFα constructs if the endogenous EGFR are first blocked with antagonistic antibodies (unpublished observations). Second, many of the studies on juxtacrine signaling have used cell types that are hypersensitive to EGFR ligands, usually because of EGFR overexpression (12–14). Even though these systems can be very useful, they may overestimate the degree of juxtacrine signaling. Finally, most studies have not quantified the relative contribution of soluble versus membrane-anchored ligand to total ligand activities. In those cases where this comparison has been done, soluble ligands have been found to have much greater activity than their membrane-anchored counterparts (12, 14, 40).
Our data indicate that in the case of autocrine signaling through the EGFR, conversion of the membrane-anchored to a soluble form is required to observe significant biological activity. Interestingly, we found that although HMEC released almost 10-fold more AR than TGFα, autocrine-stimulated mitogenesis correlated with the release of TGFα. This difference could be caused by the much lower affinity of AR for the EGFR as compared with TGFα (41). Alternately, the biological activity of AR appears to depend on the expression of specific glycosaminoglycans by target cells (42), a parameter we could not control. In any case, the tight correlation that we observed between TGFα release and proliferation of HMEC strongly indicates that the proteolytic release of EGFR ligands is the rate-limiting step in EGFR activation.
Although proteolytic release appears to be necessary for the activity of EGF and TGFα, it may not be true for all EGFR ligands. For example, good evidence exists that heparin-binding EGF can operate efficiently in a juxtacrine mode (14). However, TGFα and AR appear to be the most important ligands in EGFR-dependent tissues, such as the skin, gut, and mammary epithelium (3, 43, 44). Recently, it has been observed that proteolytic release of membrane-anchored neuregulins is necessary for their in vivo activity (45). In addition, proteolytic processing of the Notch ligand Delta also appears necessary for expression of its biological activity (46). Interestingly, the protease responsible for the processing of the Notch ligand is a member of the ADAM family of metalloproteases. A member of this family, TACE/ADAM 17, recently has been shown to be responsible for the release of TGFα (10). The ADAM proteases contain both disintegrin and signaling domains that potentially could regulate both their localization and activity (47). Recently, we have shown that the membrane-anchoring domain of EGFR ligands serves an important function in regulating the cell compartment in which ligand can access the EGFR (18). A requirement for both specific localization and regulated proteolysis of ligands could be an important determinant of EGFR activation.
Metalloprotease inhibitors that prevent EGFR ligand release, such as batimastat, have shown promise in preventing spread of metastatic disease in both animal studies and early clinical trials (22, 23). These inhibitors originally were thought to work by preventing the proteolytic breakdown of the extracellular matrix by invading tumor cells (37). Detailed studies, however, have suggested that they also inhibit cell proliferation and migration (24). EGFR activation is necessary for the growth of many epithelial tumors and can regulate the synthesis of a number of matrix-degrading proteases (48). The inhibitory effect of batimastat on metastasis thus may be mediated in part through interference with autocrine EGFR signaling. Efforts to identify proteases involved in growth factor processing together with rational drug design to selectively inhibit their activity therefore may be a fruitful approach to improve current cancer therapies. In addition, understanding how proteolysis of EGFR ligands is regulated may provide additional insights into how cell behavior is coordinated during development.
Acknowledgments
We thank Margaret Woolf and Virginia Hill for excellent technical assistance and Angie Kollhoff for providing the inspiration for these experiments. This work was supported by grants from the U.S. Army Medical Research and Materiel Command Breast Cancer Program, the National Institutes of Health, the National Science Foundation, and a merit review grant from the Veterans Administration.
ABBREVIATIONS
- AR
amphiregulin
- EGF
epidermal growth factor
- EGF-Ct
EGF with carboxyl terminus
- EGFR
EGF receptor
- HMEC
human mammary epithelial cells
- sEGF
secreted EGF
- TGFα
transforming growth factor α
References
- 1.Miettinen P J, Berger J E, Meneses J, Phung Y, Pedersen R A, Werb Z, Derynck R. Nature (London) 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- 2.Threadgill D W, Dlugosz A A, Hansen L A, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris R C, et al. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 3.Derynck R. Adv Cancer Res. 1992;58:27–52. doi: 10.1016/s0065-230x(08)60289-4. [DOI] [PubMed] [Google Scholar]
- 4.Shoyab M, Plowman G D, McDonald V L, Bradley J G, Todaro G J. Science. 1989;243:1074–1076. doi: 10.1126/science.2466334. [DOI] [PubMed] [Google Scholar]
- 5.Higashiyama S, Abraham J A, Miller J, Fiddes J C, Klagsbrun M. Science. 1991;251:936–939. doi: 10.1126/science.1840698. [DOI] [PubMed] [Google Scholar]
- 6.Shing Y, Christofori G, Hanahan D, Ono Y, Sasada R, Igarashi K, Folkman J. Science. 1993;259:1604–1607. doi: 10.1126/science.8456283. [DOI] [PubMed] [Google Scholar]
- 7.Massague J, Pandiella A. Annu Rev Biochem. 1993;62:515–541. doi: 10.1146/annurev.bi.62.070193.002503. [DOI] [PubMed] [Google Scholar]
- 8.Black R A, Rauch C T, Kozlosky C J, Peschon J J, Slack J L, Wolfson M F, Castner B J, Stocking K L, Reddy P, Srinivasan S, et al. Nature (London) 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 9.Moss M L, Jin S L, Milla M E, Bickett D M, Burkhart W, Carter H L, Chen W J, Clay W C, Didsbury J R, Hassler D, et al. Nature (London) 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 10.Peschon J J, Slack J L, Reddy P, Stocking K L, Sunnarborg S W, Lee D C, Russell W E, Castner B J, Johnson R S, Fitzner J N, et al. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 11.Sibilia M, Wagner E F. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 12.Brachmann R, Lindquist P B, Nagashima M, Kohr W, Lipari T, Napier M, Derynck R. Cell. 1989;56:691–700. doi: 10.1016/0092-8674(89)90591-6. [DOI] [PubMed] [Google Scholar]
- 13.Wong S T, Winchell L F, McCune B K, Earp H S, Teixido J, Massague J, Herman B, Lee D C. Cell. 1989;56:495–506. doi: 10.1016/0092-8674(89)90252-3. [DOI] [PubMed] [Google Scholar]
- 14.Higashiyama S, Iwamoto R, Goishi K, Raab G, Taniguchi N, Klagsbrun M, Mekada E. J Cell Biol. 1995;128:929–938. doi: 10.1083/jcb.128.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher D A, Salido E C, Barajas L. Annu Rev Physiol. 1989;51:67–80. doi: 10.1146/annurev.ph.51.030189.000435. [DOI] [PubMed] [Google Scholar]
- 16.Barnard J A, Graves-Deal R, Pittelkow M R, DuBois R, Cook P, Ramsey G W, Bishop P R, Damstrup L, Coffey R J. J Biol Chem. 1994;269:22817–22822. [PubMed] [Google Scholar]
- 17.Hashimoto K, Higashiyama S, Asada H, Hashimura E, Kobayashi T, Sudo K, Nakagawa T, Damm D, Yoshikawa K, Taniguchi N. J Biol Chem. 1994;269:20060–20066. [PubMed] [Google Scholar]
- 18.Wiley H S, Woolfe M F, Opresko L K, Burke P M, Will B H, Morgan J A, Lauffenburger D A. J Cell Biol. 1998;143:1317–1328. doi: 10.1083/jcb.143.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arribas J, Coodly L, Vollmer P, Kishimoto T K, Rose-John S, Massague J. J Biol Chem. 1996;271:11376–11382. doi: 10.1074/jbc.271.19.11376. [DOI] [PubMed] [Google Scholar]
- 20.Lanzrein M, Garred O, Olsnes S, Sandvig K. Biochem J. 1995;310:285–289. doi: 10.1042/bj3100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dempsey P J, Meise K S, Yoshitake Y, Nishikawa K, Coffey R J. J Cell Biol. 1997;138:747–758. doi: 10.1083/jcb.138.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Fu X, Brown P D, Crimmin M J, Hoffman R M. Cancer Res. 1994;54:4726–4728. [PubMed] [Google Scholar]
- 23.Parsons S L, Watson S A, Steele R J. Eur J Surg Oncol. 1997;23:526–531. doi: 10.1016/s0748-7983(97)93077-8. [DOI] [PubMed] [Google Scholar]
- 24.Chambers A F, Matrisian L M. J Natl Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 25.Stampfer M R, Pan C H, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P. Exp Cell Res. 1993;208:175–188. doi: 10.1006/excr.1993.1236. [DOI] [PubMed] [Google Scholar]
- 26.Stampfer M R, Yaswen P. Cancer Surv. 1993;18:7–34. [PubMed] [Google Scholar]
- 27.Band V, Sager R. Proc Natl Acad Sci USA. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill G N, Kawamoto T, Cochet C, Le A, Sato J D, Masui H, McLeod C, Mendelsohn J. J Biol Chem. 1984;259:7755–7760. [PubMed] [Google Scholar]
- 29.Will B H, Lauffenburger D A, Wiley H S. Tissue Eng. 1995;1:83–96. doi: 10.1089/ten.1995.1.81. [DOI] [PubMed] [Google Scholar]
- 30.Dempsey P J, Coffey R J. J Biol Chem. 1994;269:16878–16889. [PubMed] [Google Scholar]
- 31.Kamps M P, Sefton B M. Oncogene. 1988;2:305–315. [PubMed] [Google Scholar]
- 32.Dickinson R B, Tranquillo R T. AIChE J. 1993;39:1995–2010. [Google Scholar]
- 33.Coffey R J, Hawkey C J, Damstrup L, Graves-Deal R, Daniel V C, Dempsey P J, Chinery R, Kirkland S C, DuBois R N, Jetton T L, Morrow J D. Proc Natl Acad Sci USA. 1997;94:657–662. doi: 10.1073/pnas.94.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watkins L F, Levine A E. Int J Cancer. 1991;47:455–460. doi: 10.1002/ijc.2910470325. [DOI] [PubMed] [Google Scholar]
- 35.Danos O, Mulligan R C. Proc Natl Acad Sci USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fry D W, Kraker A J, McMichael A, Ambroso L A, Nelson J M, Leopold W R, Connors R W, Bridges A J. Science. 1994;265:1093–1095. doi: 10.1126/science.8066447. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen H S, McCann P P. Pharmacol Ther. 1997;75:69–75. doi: 10.1016/s0163-7258(97)00023-5. [DOI] [PubMed] [Google Scholar]
- 38.Barrandon Y, Green H. Cell. 1987;50:1131–1137. doi: 10.1016/0092-8674(87)90179-6. [DOI] [PubMed] [Google Scholar]
- 39.Matthay M A, Thiery J P, Lafont F, Stampfer F, Boyer B. J Cell Sci. 1993;106:869–878. doi: 10.1242/jcs.106.3.869. [DOI] [PubMed] [Google Scholar]
- 40.Ono M, Raab G, Lau K, Abraham J A, Klagsbrun M. J Biol Chem. 1994;269:31315–31321. [PubMed] [Google Scholar]
- 41.Adam R, Drummond D R, Solic N, Holt S J, Sharma R P, Chamberlin S G, Davies D E. Biochim Biophys Acta. 1995;1266:83–90. doi: 10.1016/0167-4889(94)00224-3. [DOI] [PubMed] [Google Scholar]
- 42.Johnson G R, Wong L. J Biol Chem. 1994;269:27149–27154. [PubMed] [Google Scholar]
- 43.Li S, Plowman G D, Buckley S D, Shipley G D. J Cell Physiol. 1992;153:103–111. doi: 10.1002/jcp.1041530114. [DOI] [PubMed] [Google Scholar]
- 44.Piepkorn M, Pittelkow M R, Cook P W. J Invest Dermatol. 1998;111:715–721. doi: 10.1046/j.1523-1747.1998.00390.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Hwang H, Cao L, Buckland M, Cunningham A, Chen J, Chien K R, Graham R M, Zhou M. Proc Natl Acad Sci USA. 1998;95:13024–13029. doi: 10.1073/pnas.95.22.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi H, Rand M D, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 47.Black R A, White J M. Curr Opin Cell Biol. 1998;10:654–659. doi: 10.1016/s0955-0674(98)80042-2. [DOI] [PubMed] [Google Scholar]
- 48.McDonnell S E, Kerr L D, Matrisian L M. Mol Cell Biol. 1990;10:4284–4293. doi: 10.1128/mcb.10.8.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]