Abstract
The database of 3D interacting domains (3did) is a collection of protein interactions for which high-resolution 3D structures are known. 3did exploits structural information to provide the crucial molecular details necessary for understanding how protein interactions occur. Besides interactions between globular domains, the new release of 3did also contains a hand-curated set of transient peptide-mediated interactions. The interactions are grouped in Interaction Types, based on the mode of binding, and the different binding interfaces used in each type are also identified and catalogued. A web-based tool to query 3did is available at http://3did.irbbarcelona.org.
INTRODUCTION
Proteins are the main perpetrators of most biological processes that take place within and between cells. However, proteins are very social in nature and often perform their function as part of large molecular machines, whose action is coordinated through complex regulatory networks of transient protein interactions. It is thus the relationships between molecules, rather than their mere presence, what will ultimately determine the behavior of a biological system. Consequently, after the completion of the first genome sequencing projects, much effort has been devoted to unveiling protein interrelationships in a high-throughput manner, and recent years have witnessed the consecution of the first interactome drafts for several model organisms, including human (1,2), setting the bases for future systems biology initiatives (3). However, high-throughput interaction discovery experiments indicate only that two proteins interact, but do not provide information about the molecular details or the mechanism of the interaction. Currently, this atomic level of detail can come only from high-resolution 3D structures, where the residue contacts are resolved and the protein interaction interfaces characterized. As a result, several databases have been developed in the last years to capture and store interactions of known 3D structure (4–6).
The database of 3D interacting domains (3did) is a collection of protein–protein interactions for which a high-resolution 3D structure has been solved. By exploring all interactions of known structure as stored in the Protein Data Bank (PDB) (7), we could divide them into two main categories on the basis of their contact interfaces: domain–domain and domain–peptide interactions (3). We also used the finding that homologous pairs of interacting proteins tend to interact in the same way (i.e. all FGFs bind the same FGF receptor pocket) to further cluster and classify protein interactions in Interaction Types (8), according to their binding and interface topologies.
DOMAIN–DOMAIN INTERACTIONS
Domain–domain interactions involve the binding of two globular domains, which creates a large contact interface of ∼2000 Å2 on average (9). These are the type of interactions that usually occur in multimeric enzymes and large multiprotein complexes, and they can be either intra- or inter-molecular (i.e. between domains in the same or different proteins, respectively).
To identify all the cases of domain–domain interactions of known 3D structure, we first assigned Pfam (10) domains to each individual protein in the PDB. We then computed all the physical interactions between domains requiring at least five contacts (hydrogen bonds, electrostatic or van der Waals interactions), and removed those lacking a significant interface as described in refs (11,12). This procedure has proven efficient at identifying and purging interaction artifacts from crystal packing; however, it is likely that 3did still contains some nonbiological associates. Currently, 3did contains 115 559 domain–domain interactions of known 3D structure comprising 120 980 proteins. We have classified them in 4887 unique interaction types according to the Pfam families mediating them. Of these, 3535 interaction types always occur between domains placed in different proteins (intermolecular), 738 are only seen between domains in the same polypeptide chain (intramolecular) and the remaining 614 occur both inter- and intra-molecular. When available, 3did also contains functional information about the interacting domains as annotated in the Gene Ontology (GO) database (13).
PEPTIDE-MEDIATED INTERACTIONS
Domain–peptide, or peptide-mediated, interactions are those where a globular domain in one protein recognizes and binds a short linear motif in another, creating a relatively small interface. Such interactions are found predominantly in signaling and regulatory networks (14) and, due to their transient nature, are much more difficult to handle biochemically. Linear motifs are short patterns of around 10 residues with a common function (i.e. binding to a globular domain) that occur in otherwise unrelated proteins. In isolation, these motifs bind their target proteins with sufficient strength to establish a functional interaction. They are frequently found in disordered or unstructured regions and adopt a well-defined structure only upon binding. An example of this type of interactions is the well-studied Src-homology-3 (SH3) domain, which binds slightly different variants of proline-rich peptides (e.g. [RKY]xxPxxP or PxxPx[KR]). Most of what is currently known about peptide-mediated interactions is compiled in the Eukaryotic Linear Motif (ELM) database (15), which provides a literature-curated collection of motifs and their interaction partners. Finally, it is worth remembering that these interactions, as the domain–domain ones, can also be intra- or inter-molecular.
Our procedure to detect all cases of peptide-mediated protein interactions of known 3D structure was recently described in ref. (16). In brief, we first parsed the PDB and identified all those entries containing two or more interacting proteins. We extracted all the information regarding the different 66 ligands involved in peptide-mediated interactions from the ELM database and assigned Pfam families to all the globular domains involved in the interactions via literature curation. We then assigned Pfam families to all interactions of known 3D structure. Whenever we identified a protein chain containing an ELM-binding domain, we searched all contacting chains for occurrences of the linear consensus motif. When we found a motif match in close vicinity of the globular domain (≤10 Å) we considered it a potential domain–peptide interaction. Finally, we went manually through the 2200 potential hits, comparing the interacting structures to those described in the literature, and removing false positives where the interaction was not mediated by the consensus peptide. Because of the visual inspection, we are confident that the interactions reported here are biologically relevant. At present, 3did contains data on 829 hand-curated peptide-mediated interactions of known 3D structure, from 611 protein pairs, involving 32 globular domains and 51 linear motifs.
IDENTIFICATION OF INTERACTION INTERFACES
The whole concept of interaction types relies on the observation that homologous pairs of interacting proteins very often interact in the same way, this is, using the same binding interfaces (8). However, there are exceptions to the norm where homologous protein pairs can interact in a completely different manner. For instance, the interaction between the signaling proteins CheY and CheA-P2 differs by a rotation of 90° in different bacterial species, despite being close homologs (17). This is particularly relevant for those proteins that have evolved to interact with many different partners by only changing a few binding residues, such as antibodies, ankyrin repeats, etc. (18).
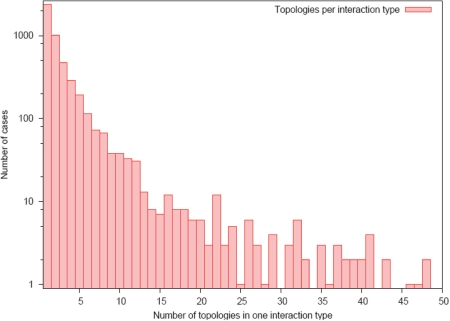
To encapsulate this information into 3did, we have computed and classified all the interaction interfaces for each interaction type using a clustering procedure reminiscent of the one used by Kim et al. (19). For each interface of a given interaction type, we identified all the contacting residues in the two domains. We then computed a distance matrix for all the interfaces based on the number of shared contacts, and performed a complete linkage hierarchical clustering analysis to discover the different modes of interaction between the two given domains. The result is that, for each interaction type, we are able to identify how many different interfaces are used and how often these occur. We have termed the alternative interaction interfaces within the same interaction type Interface Topologies, and they are stored in 3did together with the frequency in which they occur. Although the vast majority of known interaction types only display one or a few different topologies, it is also true that some families are able to interact with many partners using a large number of surface patches (Figure 1). It is thus important, if one wants to model the structure of one interaction onto another, to make sure that, for this particular interaction type, only one interaction topology is possible or, at least, that there is one whose occurrence clearly stands over the rest.
Figure 1.
Number of interface topologies per interaction type. Half of the interaction types in 3did always interact using the same topology, and most of the remaining ones show only a few different topologies. For a handful of interaction types, we find over 50 interface topologies (66 for Ras:Ras up to 199 for V-set:V-set).
3did Usage and Visualization
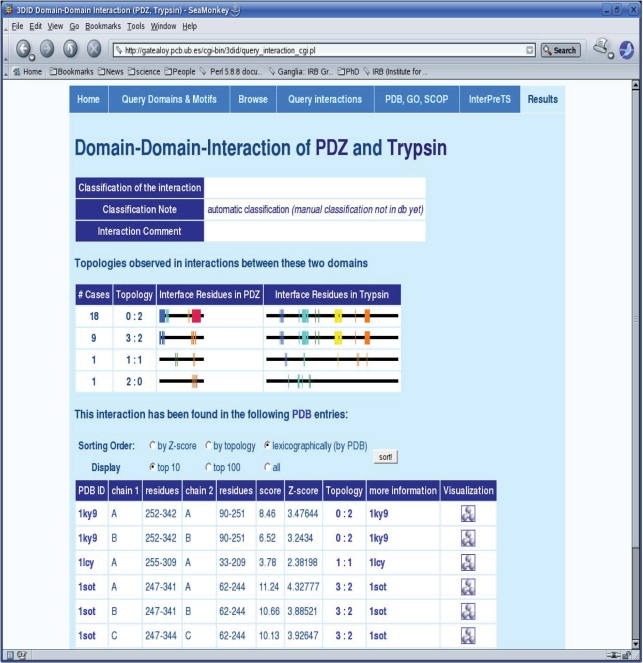
The standard way of accessing 3did is through the web-based tool by querying it with a particular domain or motif, although it can also be queried by pasting a protein sequence or directly indicating the PDB codes or GO terms of interest. As in previous versions, 3did will then display all domains, or peptides, that do physically interact with our domain of interest and for which the 3D structure of the interaction is known. All interaction partners will also be displayed in an interactive network (Figure 2), where the user can choose the depth and a color scheme based on molecular function, biological process or cellular compartment as described by GO. The network also gives information on the type of interaction (domain–domain or peptide-mediated) and whether these interactions are intra- or inter-molecular. The user can then select a particular interaction and retrieve the specific details stored in 3did. The output page for each domain–domain interaction displays a table with information concerning all the known 3D structures where this interaction is found (Figure 3). The table shows the exact location of the two domains in the 3D complex and gives empirical potential scores and Z-scores, which provide a measure of the number of favorable interacting residue-pairs at the interface (11,12). The Z-score generally accounts for interaction specificity: the higher it is the more specific the interaction. Finally, clicking on the rasmol (20) icon pops up a display of the 3D complex. The two interacting domains are colored and shown in ribbons representation with the residues participating in the interface (i.e. making hydrogen bonds, salt bridges or van der Waals contacts) shown in ball-and-stick. The newest version of 3did also includes a graphical representation of the different interaction topologies for each interacting domain. This representation indicates which residues of a domain are used in a particular interaction, as well as their frequency (Figure 3).
Figure 2.
Motif query results. The query results for ‘LIG_SH2_SRC’ show the linear motif pattern and source database (ELM), links to the binding domain SH2 and all 3D structures containing this motif, followed by all motifs binding SH2 along with their patterns (if available), SH2's interface residues and a link to the corresponding domain–motif interaction page. The network below visualizes domains and motifs interacting with LIG_SH2_SRC as well as their interactions among each other.
Figure 3.
Domain–domain interactions with interface topologies. The domain–domain interaction view shows all topologies observed in 3D structures of this interaction type along with their frequencies. The ‘rainbow’ color scheme is used to visualize where interface residues lie in the sequence, from N-terminus (blue) to C-terminus (red). Each topology has an identifier (ID) of the form ‘X : Y’, where X is the interface ID in domain 1 (PDZ here) and Y is the interface ID of domain 2 (Trypsin here). Note that for homomeric interactions, ‘X : X’ indicates a symmetric interaction. The interaction details provide PDB ID, domain positions, score and Z-score as well as the topology ID, linked to the topology visualization above, for each interaction between these two domains in a known 3D structure.
AVAILABILITY
A web-based tool to query 3did is available at http://3did.irbbarcelona.org. MySQL and flat files containing the entire database are also available through the website for independent studies. 3did is weekly updated with new 3D structures, and major updates are implemented whenever new versions of Pfam or ELM are released.
FUNDING
Spanish Ministerio de Educación y Ciencia (PSE-010000-2007-1 and BIO2007-62426) partially; 3D-Repertoire from the European Commission under FP6 contract LSHG-CT-2005-512028. Funding for open access charge: BIO2007-62426.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 2.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 3.Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 4.Stein A, Russell RB, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 6.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein-protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aloy P, Russell RB. Ten thousand interactions for the molecular biologist. Nat. Biotechnol. 2004;22:1317–1321. doi: 10.1038/nbt1018. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 10.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc. Natl Acad. Sci. USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aloy P, Russell RB. InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics. 2003;19:161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 13.The Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 15.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein A, Aloy P. Contextual specificity in peptide-mediated protein interactions. PLoS ONE. 2008;3:e2524. doi: 10.1371/journal.pone.0002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SY, Beel BD, Simon MI, Bilwes AM, Crane BR. In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc. Natl Acad. Sci. USA. 2004;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J. Mol. Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein-protein interactions: a compendium of interface geometry. PLoS Comput. Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]