Abstract
Plants produce small cysteine-rich antimicrobial peptides as an innate defense against pathogens. Based on amino acid sequence homology, these peptides were classified mostly as α-defensins, thionins, lipid transfer proteins, cyclotides, snakins and hevein-like. Although many antimicrobial plant peptides are now well characterized, much information is still missing or is unavailable to potential users. The compilation of such information in one centralized resource, such as a database would therefore facilitate the study of the potential these peptide structures represent, for example, as alternatives in response to increasing antibiotic resistance or for increasing plant resistance to pathogens by genetic engineering. To achieve this goal, we developed a new database, PhytAMP, which contains valuable information on antimicrobial plant peptides, including taxonomic, microbiological and physicochemical data. Information is very easy to extract from this database and allows rapid prediction of structure/function relationships and target organisms and hence better exploitation of plant peptide biological activities in both the pharmaceutical and agricultural sectors. PhytAMP may be accessed free of charge at http://phytamp.pfba-lab.org.
INTRODUCTION
The first antimicrobial peptide from a eukaryotic organism, wheat α-purothionin, was discovered in 1942 by Balls and collaborators (1). The next peptide in this category was not reported until 30 years later and studies describing the discovery of new antimicrobial peptides from plant tissues have become numerous only in recent years (2). Antimicrobial peptides (AMPs) are cysteine-rich short amino acid sequences common in the seeds of many species (3). Plant AMPs are grouped into several families and many share general features, such as an overall positive charge, the presence of disulfide bonds (which stabilize the structure) and a mechanism of action targeting outer membrane structures, such as ion channels. In addition to their role in host defense and their appeal as simple models for studying the molecular mechanism of antimicrobial peptide action, AMPs have the potential to combat pathogens, including those showing increased resistance to conventional antimicrobial compounds. These peptides usually have broad-spectrum antimicrobial activity against pathogenic fungi and thus are promising candidates for managing diseases in sensitive transgenic plants (4). Although many plant AMPs are now well characterized, much physicochemical and structural information is still missing, unavailable to potential users or buried in the scientific literature. The majority of sequenced AMPs are stored in the manually annotated UniProtKB/Swiss-Prot which represents a large database with broad domains. Thus, there is a clear need to gather, filter and critically evaluate this mass of information and store into smaller, more specialized, resources so that it can then be used in a way that enhances efficiency. Few different databases have been created for antimicrobial peptides and are mentioned in the literature. ANTIMIC (5) database is currently inactive. The Antimicrobial Peptide Database (APD) (6) contains general information about peptides of all types having antibacterial, antifungal or antiviral activities and originating from either eukaryotic or prokaryotic cells. Plant AMPs are not described with sufficient details in this database. A centralized resource, such as a database designed specifically for plant AMPs would facilitate the comprehensive investigation of their structure/activity associations and potential uses. This could have implications not only for the genetic improvement of plants by increased resistance to pathogens, but also for the development of new drugs for medical use.
CONSTRUCTION AND CONTENT
Database construction and methods
PhytAMP runs on a Windows NT platform (Microsoft Windows 2000) with the Apache web server (version 2.0.54), MySQL server (v 5.0.30) and PHP (v 4.3.11). The web server and all parts of the database are hosted at the Centre de Calcul El Khawarizmi (CCK), Tunisia. Antimicrobial plant peptide sequences were collected from the UniProt database (7) and from the scientific literature using PubMed. Microbiological information was collected from the literature by PubMed search. Since not all known AMPs sequences were present in the ExPASy (http://www.expasy.org/srs/) SRS server or NCBI server (http://www.ncbi.nlm.nih.gov/entrez/), literature search was used to complete the PhytAMP sequence database. Sequences were retrieved in SciDBMaker (8) and curated and the resulting tables exported to the MySQL server. The FASTA program (9) was used for the sequence homology search in the database. The BLAST search (10) was implemented using the NCBI binaries. The Smith–Waterman search was implemented using the SSEARCH program from the FASTA3 distribution (9). The sequence alignment was done using various methods, such as ClustalW (v2.07) (11), MUSCLE (v3.6) (12) and T-Coffee (v1.37) (13). The Java platform is required for visualizing generated phylogenic trees. The program HMMER was used for the implementation of hidden-profile Markov models (14). The peptides collected in this version of PhytAMP are mainly from natural sources. Precursor sequences were removed to keep only mature peptide sequences. For each peptide, a unique nine-digit identification number (ID) starting with the prefix PHYT was assigned. Each entry was checked in the Protein Data Bank (PDB) or UniProtKB/Swiss-Prot. A web link in PhytAMP to UniProt and PDB was created for all peptides that already exist in these databases, to facilitate consultation of the original databases. In addition, each entry contains general data, such as peptide name, sequence, class, plant taxonomy, activity data (bacterial, fungal or viral target) and relevant references in the UniProt. Additional physicochemical data are provided, including empirical formula, mass, length, isoelectric point, net charge, the numbers of basic, acidic, hydrophobic and polar residues, hydropathy index, binding potential index, instability index, aliphatic index, half-life in mammalian cells, yeast and Escherichia coli, cysteine and glycine content, extinction coefficient, absorbance at 280 nm, absent and most prevalent amino acids, secondary (α-helix or β-strand) and tertiary structure (when available), physical method used for structural determination (e.g. NMR spectroscopy or X-ray diffraction) and critical residues for activity, when information is available.
Web interface description
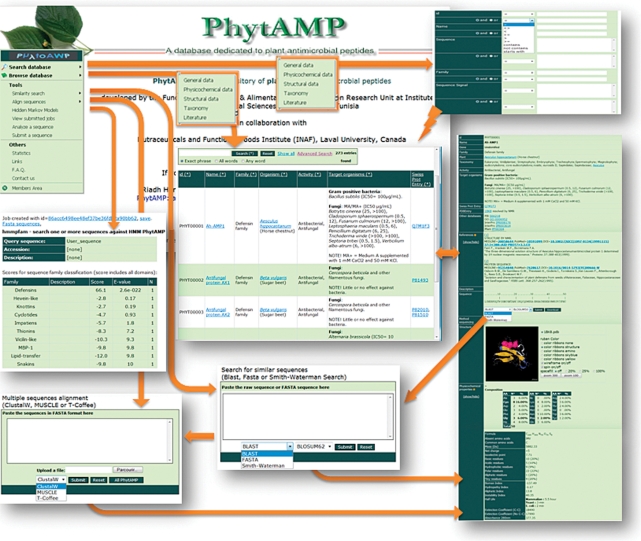
PhytAMP database is available at http://phytamp.pfba-lab.org. There are various ways to access to information related to a given peptide in PhytAMP database. The simplest way is to use the browse interfaces (general information, physicochemical data, structural data, taxonomy and literature). A quick search formula on the header part on ‘browse’ web pages is included for keyword search. An extended search interface (query for general information, physicochemical data, structural data, taxonomy and literature) is provided for combined search. Various tools and links are also provided including user sequence analysis interface, user sequence similarity search interface, statistical data, useful links and contact information (Figure 1). The query forms provide quick or advanced search with a variety of parameters. Users can find a specific antimicrobial plant peptide using its ID, name or UniProt ID, query for lists of organisms targeted by a plant AMP or for lists of AMPs that target a specific organism. Detailed information for each entry in the database can be viewed by clicking on the peptide name. The advanced search tool allows query of all available data. When a sequence is entered, the program returns all peptides containing this sequence and search results can be sorted into visible columns. A combinatorial search can be done by query of search results. Files containing the sequence (Fasta format) may be downloaded for all of the entries identified by the query, to facilitate other analyses. Registered users can also download output result tables in XLS, DOC, XML and CSV format. In addition, various tools including BLAST, FASTA and SSEARCH enable users to search the database for homologous sequences and save successful results temporarily in the server for subsequent access. Users may thus select some or all of the homologous sequences for multiple aligning with their submitted sequences. The statistical interface provides data on peptide sequence, function and structure. The average length, net charge and amino acid residue percentages for all entries in the database are also listed, as is the frequency of given values for each physicochemical parameter. For structural analysis, the number of peptides with a defined structural type is shown.
Figure 1.
User interface of PhytAMP database.
UTILITY AND DISCUSSION
Phylogenetic tree construction
Multiple sequence alignments of 271 plant antimicrobial peptides found in the PhytAMP were made using the CLUSTALW v2.07 program (11) and further refined manually. The parameters used in the CLUSTALW program were as follows: gap opening, 10; gap extension, 0.2; delay divergent sequence, 30%; DNA transition weight, 0.5; protein weight matrix, Gonnet series. Based on the initial alignment, a resample was performed by the generation of 1000 bootstrapped data sets using the SEQBOOT program (15). Genetic distances of the alignments were calculated using the Dayhoff PAM matrix with the PROTDIST program (15). Subsequently, the trees were constructed by successive clustering of lineages using the neighbor-joining algorithm as implemented in the NEIGHBOR program (15). Their strict consensus tree was obtained using the CONSENSE program (15). The unrooted tree diagram was generated with the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/). 3D structure data were obtained from the PDB (http://www.rcsb.org/pdb) and edited with the molecule analysis and molecule display (PyMOL) program (http://www.pymol.org).
The PhytAMP database
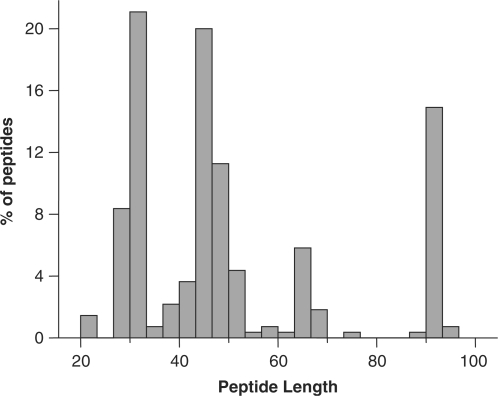
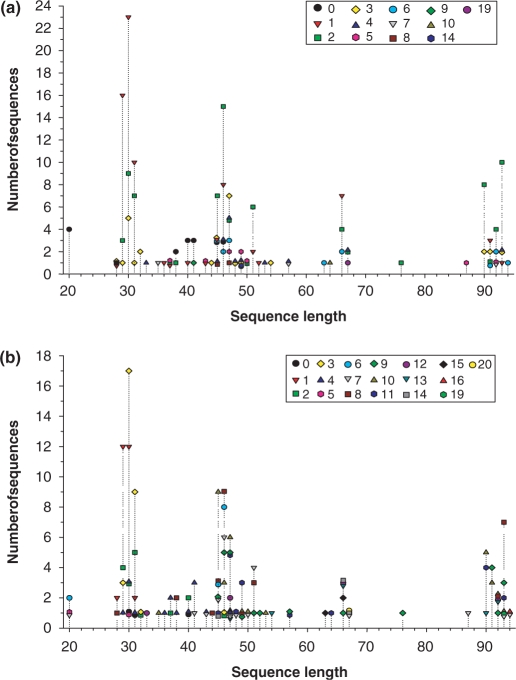
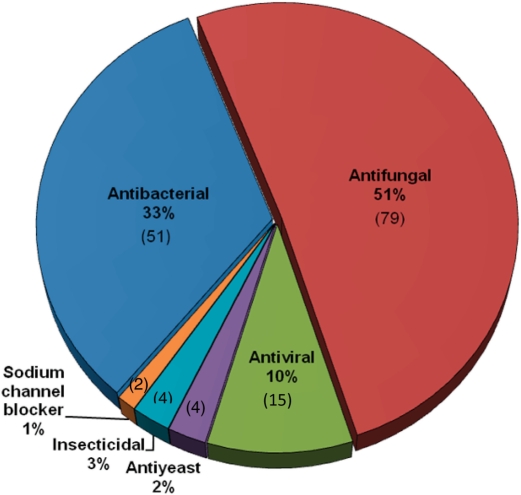
The current version of PhytAMP holds 271 antimicrobial plant peptides (AMPs), secreted by various families, such as Amaranthaceae [9], Andropogoneae [10], Brassicaceae [36], Oryzeae [11], Santalaceae [11], Spermacoceae [17], Triticeae [34], Vicieae [12] and Violaceae [51]. Classification has been proposed on the basis of primary structure (16, 17). Viola (family Violaceae) and Arabidopsis (family Brassicaceae) appear to be the predominant genera among AMP producers, although this may be due to the extensive studies on these species. Plant AMPs in the database are classified as cyclotides [76], defensins [55], Hevein-like [14], Impatiens [4], knottins [4], lipid-transfer proteins [45], shepherins [2], snakins [20], thionins [43] or vicilin-like [6], MBP-1 (18) and beta-barellin (19). An unrooted tree of the AMPs was generated, as shown in Figure 2. It is noteworthy that only 69% of the peptides have been sequenced directly, the remaining structures having been predicted from genome sequences. For 83.4%, the amino acid sequence length varies from 20–67 (Figure 3). Table 1 summarizes the amino acid percentages. It is generally presumed that AMPs are cysteine-rich proteins and this was apparent in our statistical results. Glycine is also an abundant amino acid, 98.5% of these AMPs containing at least one glycine residue. The majority (84.9%) have net charges varying from 0 to +10, while <6% possess a positive charge superior to +10, the highest being +17 (PHYT00099). In addition, only 9.2% have a net negative charge, the most negative being −6 (PHYT00259). As a result, the average net charge of all AMPs in PhytAMP is +4.6. Figure 4 shows the correlation between acidic (a) and basic (b) amino acid content and sequence length among peptides in the PhytAMP database. In general, peptides are randomly distributed across families, except sequence length 20 which correspond to cyclotide family and the cluster for sequence length about 90 which fall specifically in lipid-transfer protein family. The majority of sequences display a basic pattern, 53.1% having from 6–11 basic residues. In comparison, acidic residue content is more limited, 79.7% containing three or fewer acidic amino acids. Current analysis revealed that three quarters of the plant AMPs contain between 4 and 13 hydrophobic residues. Only 39 were found to have 3D structures filed in the PDB database and resolved by NMR spectroscopy, crystallography or molecular modeling. Some of them nevertheless possess more than one structure in the PDB database, bringing the total number of 3D entries to 102. Only 39.5% are tested for biological activity. The majority possesses antifungal (51%), antibacterial (33%) and antiviral (10%) activities, as shown in Figure 5. These findings may be useful in isolating and characterizing novel plant AMPs or designing novel peptides with higher potency against pathogens or with broad antimicrobial spectra. As future development, we plan to integrate a system that will allow automatic prediction of the amino acids that are key to biological function and a server for building tertiary structures by homology with existent plant AMP structures.
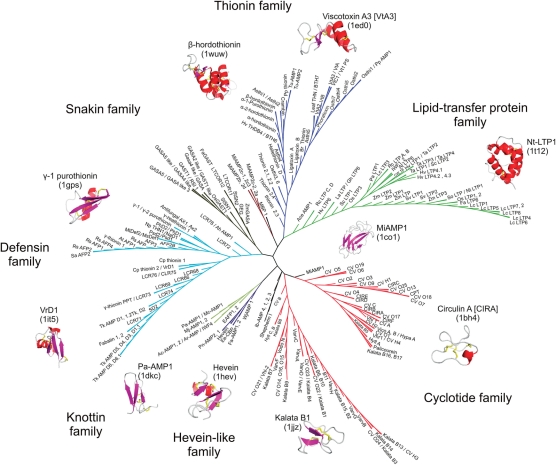
Figure 2.
Unrooted phylogenetic tree of plant AMPs compiled in the PhytAMP database. A multiple sequence alignment of 271 plant AMPs was used to calculate a matrix with the genetic distances for each pair of the sequences. Based on this matrix, successive clustering of lineages was done to construct the unrooted tree with the neighbor-joining algorithm (8). Tree was generated using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). 3D coordinates were obtained from the PDB (http://www.rcsb.org/pdb/). PDB accession ID numbers:—Viscotoxin A3: 1ed0; β-hordothionin: 1wuw; Nt-LTP1: 1t12; MiAMP1: 1co1; Circulin A: 1bh4; Kalata B1: 1jjz; Hevein: 1hev; Pa-AMP1: 1dkc; VrD1: 1it5; γ-1-purothionin: 1gps. Pictures were generated using PyMOL software [10]. α-helices and β-sheets are shown in red and purple, respectively.
Figure 3.
Histogram of peptide length distribution in the PhytAMP database.
Table 1.
Amino acid occurrence in the PhytAMP database
| Amino acid | Number of residues | % of total residues |
|---|---|---|
| C (cysteine) | 1975 | 14.59 |
| G (glycine) | 1384 | 10.23 |
| S (serine) | 1185 | 8.76 |
| A (alanine) | 979 | 7.23 |
| K (lysine) | 896 | 6.62 |
| T (threonine) | 853 | 6.30 |
| R (arginine) | 819 | 6.05 |
| P (proline) | 817 | 6.04 |
| N (asparagine) | 743 | 5.49 |
| V (valine) | 610 | 4.51 |
| I (isoleucine) | 562 | 4.15 |
| L (leucine) | 549 | 4.06 |
| Y (tyrosine) | 414 | 3.06 |
| Q (glutamine) | 410 | 3.03 |
| D (aspartic acid) | 335 | 2.48 |
| E (glutamic acid) | 322 | 2.38 |
| F (phenylalanine) | 291 | 2.15 |
| H (histidine) | 194 | 1.43 |
| W (tryptophan) | 123 | 0.91 |
| M (methionine) | 73 | 0.54 |
Figure 4.
Correlation between acidic (a) and basic (b) amino acid content and sequence length among peptides in the PhytAMP database.
Figure 5.
Chart of reported activities for plant peptides compiled in the PhytAMP database.
CONCLUSION
PhytAMP allows all plant AMP sequence information and physicochemical or biological data to be accessed via a user-friendly, web-based interface. The database can be queried using various criteria and retrieval of microbiological or physicochemical data includes specific information on each peptide. The microbiological, physicochemical and structural proprieties thus provided should allow more comprehensive analysis of this group of antimicrobial peptides and enhance our understanding of plant defense biology. This could contribute not only to genetic improvement of plants by increased resistance to pathogens, but also has implications for the development of new drugs for medical use based on derivatives or analogs of natural antimicrobial peptides. PhytAMP currently contains 271 entries of plant AMPs and is expected to grow quickly with the rapid development of genomic and proteomic projects. As more information about plant AMPs becomes available, the database will be expanded and improved accordingly.
FUNDING
The Natural Sciences and Engineering Research Council of Canada; the Ministry of Higher Education, Scientific Research and Technology, Republic of Tunisia.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Authors thank Dr. Stephen Davids for his critical reading of the article.
REFERENCES
- 1.Balls AK, Hale WS, Harris TH. A crystalline protein obtained from a lipoprotein of wheat flour. Cereal Chem. 1942;19:279–288. [Google Scholar]
- 2.Broekaert WF, Terras FR, Cammue BP, Osborn RW. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995;108:353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broekaert WF, Cammue BPA, De Bolle MFC, Thevissen K, De Samblanx GW, Osborn RW. Antimicrobial peptides from plants. Critical Rev. Plant Sci. 1997;16:297–323. [Google Scholar]
- 4.Terras FR, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J, et al. Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmachary M, Krishnan SPT, Koh JLY, Khan AM, Seah SH, Tan TW, Brusic V, Bajic VB. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004;32:D586–D589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Wang G. APD: the Antimicrobial Peptide Database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammami R, Zouhir A, Naghmouchi K, Ben Hamida J, Fliss I. SciDBMaker: new software for computer-aided design of specialized biological databases. BMC Bioinformatics. 2008;9:121. doi: 10.1186/1471-2105-9-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 14.Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press: Cambridge; 1998. [Google Scholar]
- 15.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Garcia-Olmedo F, Molina A, Alamillo JM, Rodriguez-Palenzuela P. Plant defense peptides. Biopolymers. 1998;47:479–491. doi: 10.1002/(SICI)1097-0282(1998)47:6<479::AID-BIP6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 17.Castro MS, Fontes W. Plant defense and antimicrobial peptides. Protein Pept. Lett. 2005;12:13–8. [PubMed] [Google Scholar]
- 18.Duvick JP, Rood T, Rao AG, Marshak DR. Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J. Biol. Chem. 1992;267:18814–18820. [PubMed] [Google Scholar]
- 19.McManus AM, Nielsen KJ, Marcus JP, Harrison SJ, Green JL, Manners JM, Craik DJ. MiAMP1, a novel protein from Macadamia integrifolia adopts a Greek key beta-barrel fold unique amongst plant antimicrobial proteins. J. Mol. Biol. 1999;293:629–638. doi: 10.1006/jmbi.1999.3163. [DOI] [PubMed] [Google Scholar]