Abstract
The TcSNP database (http://snps.tcruzi.org) integrates information on genetic variation (polymorphisms and mutations) for different stocks, strains and isolates of Trypanosoma cruzi, the causative agent of Chagas disease. The database incorporates sequences (genes from the T. cruzi reference genome, mRNAs, ESTs and genomic sequences); multiple sequence alignments obtained from these sequences; and single-nucleotide polymorphisms and small indels identified by scanning these multiple sequence alignments. Information in TcSNP can be readily interrogated to arrive at gene sets, or SNP sets of interest based on a number of attributes. Sequence similarity searches using BLAST are also supported. This first release of TcSNP contains nearly 170 000 high-confidence candidate SNPs, derived from the analysis of annotated coding sequences. As new sequence data become available, TcSNP will incorporate these data, mapping new candidate SNPs onto the reference genome sequences.
INTRODUCTION
Trypanosoma cruzi is a protozoan pathogen that infects humans and other mammals, producing a pathology called Chagas disease. The disease is endemic in most of central and South America affecting ∼18 million people (1), with an increasing number of cases in North America (2).
Trypanosoma cruzi is diploid, with a predominantly clonal (asexual) mode of replication (3), and a high degree of sequence and karyotype variability between strains (4). Based on a number of genetic and isoenzyme markers, the population of T. cruzi has been divided into six discrete evolutionary lineages (5,6), with other studies suggesting the existence of further genetic divisions (7,8). Infection with the parasite results in a number of pathologies and clinical outcomes—megacolon, megaesophagus and cardiomyopathy, among others—that are thought to be the result of a complex interplay between the host genetic background and the genetic variability present in the parasite population (9). Available studies in model organisms support this hypothesis (10), stressing the need for expanding our knowledge of the genetic variation present in the parasite.
The genome of T. cruzi was sequenced by a whole-genome sequencing approach, from a hybrid strain (CL Brener) composed of two divergent parental haplotypes (11,12). This choice of strain and sequencing strategy resulted in a high sequence coverage from the two parental haplotypes. Because of the high allelic variation, ∼30 Mb of sequence (out of the estimated 100 Mb of diploid genome size) were found to be present twice in the assembly (12). We have used the genome sequence information, together with sequences from various strains of T. cruzi available in public databases to map polymorphic sites present in coding sequence loci in T. cruzi. These candidate SNPs have been analyzed and characterized based on a number of attributes (allele frequency, effect on the encoded protein product, probability of being a true polymorphism, their overlaps with restriction enzyme sites, etc.). All this information has been integrated into a database, called TcSNP and available online at http://snps.tcruzi.org. The data integrated in this database represent the first genome-wide compilation of genetic variation data for T. cruzi. In this article, we describe the TcSNP database, the underlying data and website functionality and demonstrate its application in a number of case scenarios.
OVERVIEW OF THE TCSNP DATABASE
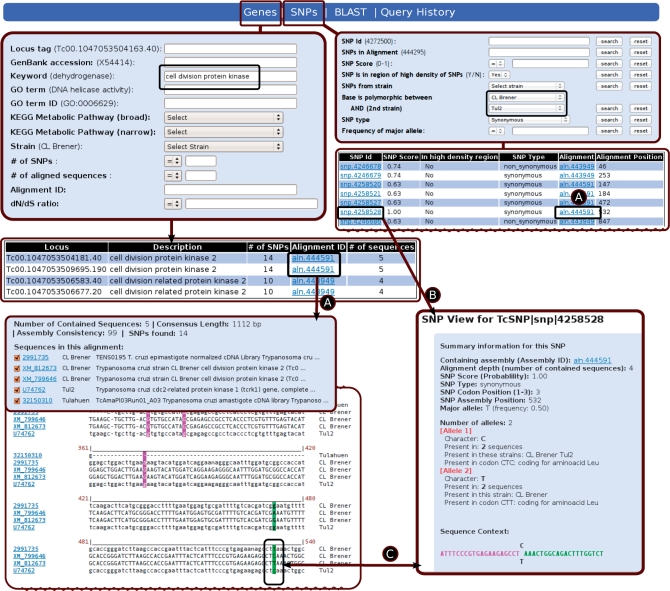
The TcSNP database contains T. cruzi sequences, multiple sequence alignments obtained from these sequences, and single-nucleotide polymorphisms and small indels identified by scanning these multiple sequence alignments (Table 1). Interrogation of the data available in TcSNP can be performed on attributes from each of these objects (Figure 1). For sequences, the database offers text-based searches on attributes derived from their annotation, such as: gene names, locus and database identifiers, gene ontology terms (molecular function, cellular process and components), biochemical pathways, strain from which the gene has been sequenced, etc. In any of these cases, the result is a list of genes matching the specified criteria, containing links to the corresponding multiple sequence alignments, where users can visualize polymorphic sites in different colors, typefaces and styles, as an indication of SNP probability and the effect on the encoded protein sequence (Figure 1). Any result set containing genes can be used to obtain the corresponding set of polymorphic sites present in those genes.
Table 1.
Summary of data available in the current release of TcSNP, showing the numbers of sequences, alignments and SNPs
| Sequences | Number | Strains |
|---|---|---|
| Reference coding sequencesa | 25 013 | 1 |
| Expressed sequence tags | 13 968 | 3 |
| Other (mRNAs, genomic) | 2038 | 295b |
| Alignments | ||
| Total No. of alignments | 7482 | |
| Alignments with two reference sequencesc | 5280 | |
| SNPsd | ||
| Total No. of SNPs | 269 686 | |
| With P > 0.7 | 195 160 | |
| Within high quality neighborhoodse | 204 823 | |
| Synonymous | 110 031 | |
| Non-synonymous | 111 117 |
aFrom the reference CL Brener genome (12).
bThis figure includes redundancy in strain names, see Methods section for more information.
cAllelic variants of the two CL Brener haplotypes.
dNumber of SNPs in each row is independent from other rows.
eLess than three SNPs in a window of 10 bp.
Figure 1.
Example search session showing the navigation flow in the TcSNP website. Users can do a gene-centric search (e.g. using the keywords ‘cell division protein kinase’), a SNP-centric search (e.g. SNP is polymorphic between strains Tul2 and CL Brener) or a sequence similarity-based search (using BLAST, not shown). From any list of results users can access the corresponding multiple sequence alignment of interest (path A), and view SNP-specific information (e.g. quality score, mutation type, detected alelles, etc.) (paths B and C).
The polymorphic sites available in TcSNP have been characterized based on a number of criteria. Using PolyBayes (13), we have calculated the probability of these sites being true polymorphic sites (as opposed to sequencing errors, see Methods section). Also, we have obtained a measure of the quality of the sequence around each site by noting the distance between neighboring polymorphic sites. Based on this analysis, we have marked sites that are located inside (and outside) of sequence regions containing three or more polymorphic sites in a window of 10 bp. Finally, we have assessed the change introduced by each putative SNP on the encoded protein (either synonymous, non-synonymous substitutions or premature stops), and provide the ratio of dN (number of nonsynonymous changes per nonsynonymous site) and dS (number of synonymous changes per synonymous site) values for a significant portion of the alignments, as an estimate of the selection pressure acting on these genes (14). All these attributes can be used to filter SNPs and arrive at SNP sets of interest. Also, users interested in a specific genomic region can also look for SNPs using genome contig identifiers and base coordinates.
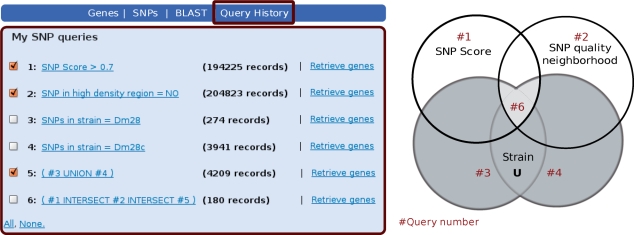
In TcSNP, all searches in a user's session are shown in the query history page, where they can be combined using standard operators (UNION, INTERSECTION and SUBTRACTION). As an example, users interested in high-confidence polymorphic sites in the T. cruzi strain Dm28 (that belongs to the evolutionary lineage Tcruzi I) can arrive at this set by asking for the INTERSECTION of SNP sets obtained for each individual criterion. This is illustrated in Figure 2 that also shows how users can overcome the existing redundancy in strain names by obtaining a UNION of SNP sets with similar strain names (see discussion about this issue in the Data sources section). As was the case for genes, SNP sets can be easily converted into the corresponding set of genes containing these SNPs.
Figure 2.
Using the query history in TcSNP to combine queries. In this example, in order to obtain high quality SNPs in a strain of interest (high score, located in good quality neighborhoods), users combine SNP sets that were obtained by filtering SNPs based on specific attributes. In the figure, the intersection of the SNP sets #1, #2 and #5 has been calculated, resulting in SNP set #6. In particular, note that #5 is the result of a union of sets #3 and #4, showing how to overcome the redundancy in strain names (Dm28c is presumably a cloned stock derived from strain Dm28). Selected queries can be combined using standard set theory operators (UNION, INTERSECTION and SUBTRACTION). On the right, a Venn diagram illustrates the operations performed on the SNP sets in this example.
In TcSNP both genes and SNPs are linked to a multiple sequence alignment, which is the source object from which all SNPs and indels were identified. Many properties that are specific for multiple sequence alignments are also searchable, such as the number of sequences contained in the alignment, the number of reference sequences from the CL-Brener genome, the number of SNPs identified in the alignment and the quality of the alignment, as estimated by two different parameters.
Users can also perform searches based on sequence similarity, by interrogating the TcSNP database with their own query sequences using the BLAST search tool integrated into the website. Currently these searches are performed against the consensus sequences of all the alignments in TcSNP.
Finally, the website provides linkouts to other web resources where users can find additional information on their genes of interest. Genes are linked to the corresponding gene pages at TcruziDB (15), GeneDB (16), TDR Targets (17) and to the corresponding source records at the NCBI.
APPLICATIONS OF THE TCSNP DATABASE
Individual researchers working with T. cruzi and/or Chagas disease are interested in sequence polymorphisms for different reasons. Molecular biologists may need information about SNPs in their genes of interest to avoid these polymorphic sites when designing oligonucleotide primers or gene knockout vectors. In contrast, researchers interested in the evolution of different T. cruzi lineages, or in the development of new typing assays might be interested in using selected SNPs as genetic markers. Genetic variation data are also important when assessing the potential for development of resistance of established drug targets; and for prioritizing candidate drug targets.
The overall functionality of the TcSNP database (available search attributes, display of multiple sequence alignments and SNPs) has been designed based on the consideration of these possible uses. The exercise illustrated in Figure 2 shows how a user can quickly arrive at genetic markers of interest. In this example, we show how to select high-quality SNPs (high SNP score, with good sequence quality neighborhoods) that are polymorphic in a strain from the evolutionary lineage Tcruzi I, and which are therefore good candidates for a diagnostic assay. Another exercise facilitated by the database is the selection of genes that are under purifying selection (dN/dS ≪ 1). This set of genes is a good starting point to look for potential drug and/or vaccine candidates. Conversely, genes under diversifying selection (dN/dS ≥ 1) might represent interesting candidates for discriminating diagnostics.
CONCLUSIONS
The TcSNP database currently represents a comprehensive resource on T. cruzi coding single nucleotide polymorphisms. In this first release of TcSNP, the dataset contains minimal information on SNPs located in intergenic (noncoding) regions of the genome (most of these SNPs are derived from the untranslated regions of ESTs, and from intergenic regions present in sequences obtained from GenBank). As expected due to the high sequence coverage for the two parental haplotypes of the CL Brener strain, and the currently limited sequence information available for other strains, the majority of these candidate SNPs correspond to heterozygous sites in CL Brener.
When interpreting SNP data in T. cruzi, the draft nature of the reference genome and its repetitive nature have to be taken into account. Sequence variation present in genes from large gene families might be underestimated in TcSNP when looking at a single multiple sequence alignment, because these families are usually represented by more than one multiple sequence alignment in the database. Although many problematic alignments have been manually curated (e.g. to join two alignments containing sequences from the same gene), the emphasis of this curation has been placed on alignments of single copy genes, where there is a low chance of dealing with paralogs. In this respect, the recent observation made by Arner et al. (18) about the collapsing of many gene copies in the genome assembly further reinforces the importance of interpreting SNP data with care. Their analysis shows that especially in the case of repetitive genes, copy numbers might have been underestimated (18). For SNP discovery, this collapsing of sequences during genome assembly, may result in an underestimation of polymorphic sites.
FUTURE WORK
Future releases of TcSNP will integrate new T. cruzi sequences as they become available. Many of these are expected to come from a resequencing effort that is underway in our lab. Planned updates to the website functionality include the development of a standardized API and web services through which other databases can consume the information provided by TcSNP and the development of functionality to design primers by interfacing with primer3 (19), while using the SNP information in this process. We encourage users to send feedback on desired features for improving the TcSNP database.
METHODS
Database and web application
The TcSNP database is composed of a web application written in Perl, running against a PostgreSQL database. The database schema is based on the Genomics Unified Schema (GUS, http://gusdb.org) with local customizations. The web application has been developed using a Model-View-Controller architectural pattern, where the work of each layer is performed by a specialized Perl component [all components are available from CPAN (20)].
Access to the database from Perl (i.e. the Model) is done through a hybrid combination of (i) an abstraction of the database schema using Perl's DBIx::Class package and (ii) custom SQL queries executed using Perl's database access interface (DBI). The controller component managing user's requests and dispatching calls to other components is Perl's Catalyst (21). A number of custom controller modules were developed, which contain the business logic of the TcSNP application. Finally, the Viewer component is Perl's Template Toolkit, which uses custom templates to render web pages using information provided by the controller. The database runs on dedicated FreeBSD servers, with the Catalyst web application running under the Apache web server.
Data sources
The reference genome sequence of the CL Brener strain of T. cruzi was obtained from GenBank using the umbrella accession number AAHK00000000 (July, 2005). Other T. cruzi sequences (mRNAs and ESTs) were also obtained from GenBank using custom Entrez queries (May, 2007). Before loading into the database, some curation has been done to standardize the names of T. cruzi strains. This was necessary because of the variations in how different authors write the names of strains in GenBank submissions and publications. For example, the ‘CL Brener’ strain appears in different GenBank sequences as CL Brener, CL-Brener, CLBrener, CL-Brenner, CL Brenner, Cl Brenner, or Cl Brener (note the different capitalization, use of middle dash, spaces and the writing of the strain name using a single or a double ‘n’). Because the database allows users to perform searches using strain names, it was important to reduce redundancy where possible. In some cases, however, it was important to keep the distinction between similar strain names, for example to discriminate between cloned stocks and their parental uncloned strains. As an example, we have kept CanIII and CanIII CL1 (CL1 stands for ‘clone 1’), and Sc43 and Sc43 CL1 as different ‘strains’ in TcSNP. Redundancy is still present, however, and further curation of strain names can be done. We also encourage users of the database to send feedback about this issue.
Sequence clustering and alignment
Before clustering all sequences were masked against a library of vector sequences and T. cruzi repetitive elements, as described previously (22). Annotated coding sequences from the reference genome, and other publicly available sequences were mapped against the genome scaffolds using BLAT. Sequences mapping to the same genomic regions were clustered together and multiple sequence alignments were obtained using phrap. This initial clustering allowed us to group mRNAs, ESTs and reference coding sequences to the reference genome assembly sequences. But because allelic variants in the CL-Brener genome were separated during assembly (12), those initial clusters showed many instances of allelic variants separated into different alignments (i.e. each mapping to its own contig). To obtain alignments between allelic variants, we merged alignments with highly similar consensus sequences (by BLAST analysis). Afterwards, and based on user feedback, we have also merged, splitted and re-analyzed many alignments. This manual curation effort was mainly focused on single copy genes. Users of TcSNP should also be aware of the fact that many sequences from the CL-Brener genome assembly are located in contigs with assembly problems or may represent assembly artifacts. For sequences containing assembly warnings in the original GenBank records, we have attached similar notes to the corresponding alignments in TcSNP to help users in the interpretation of the SNP data.
Candidate SNP identification and analysis
Multiple sequence alignments were scanned to identify polymorphic columns. To calculate the probability of these sites being true polymorphisms as opposed to sequencing errors, we have used the software package PolyBayes, version 5 (13). PolyBayes uses a Bayesian statistical framework that relies on allele frequency, alignment depth and base quality values amongst other attributes to calculate a probability score. Because chromatogram trace data are not available for many of the sequences in this release, we have devised a scoring strategy that uses arbitrary base quality values. These quality values are different depending on the sequence origin/type. Sequence bases obtained from the T. cruzi CL-Brener genome (∼19X shotgun coverage) were arbitrarily assigned a base quality value of 40; those from GenBank records, a value of 30 (individual submissions); and those from dbEST, a value of 20 (single-pass, unedited). As an example, using this scoring scheme, a single base from an EST differing from two allelic variants of CL-Brener reference sequence (depth = 3) would give a probability of 0.22 of being a true SNP (see for example: http://snps.tcruzi.org/snps/view/4028216).
To analyze the effect of each SNP on the corresponding protein product, we noted the codon position of the SNP in each reference coding sequence and evaluated the change introduced by the polymorphic base. Also, for a subset of the alignments (those containing coding sequences of similar length, with indels being a multiple of 3), we calculated dN and dS values (14) using BioPerl's population genetics modules (23).
FUNDING
National Agency for the Promotion of Science and Technology (ANPCyT, Argentina) (grant PICT 38209); and the University of San Martín (grant S05/22). Funding for open access charge: ANPCyT.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to acknowledge Patricio Diosque, Leonardo Panunzi and Raúl Cosentino for helpful comments and suggestions on the functionality of the database, and for testing early versions before release. Alejandro Ackermann is a fellow and Fernán Agüero is a member of the Research Career of the National Research Council (CONICET, Argentina).
REFERENCES
- 1.Barrett MP, Burchmore RJS, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. The trypanosomiases. Lancet. 2003;362:1469–1480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 2.Hotez PJ. Neglected infections of poverty in the United States of America. PLoS Negl. Trop. Dis. 2008;2:e256. doi: 10.1371/journal.pntd.0000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tibayrenc M, Ayala FJ. Evolutionary genetics of Trypanosoma and Leishmania. Microbes Infect. 1999;1:465–472. doi: 10.1016/s1286-4579(99)80050-1. [DOI] [PubMed] [Google Scholar]
- 4.Henriksson J, Dujardin JC, Barnabé C, Brisse S, Timperman G, Venegas J, Pettersson U, Tibayrenc M, Solari A. Chromosomal size variation in Trypanosoma cruzi is mainly progressive and is evolutionarily informative. Parasitology. 2002;124:277–286. doi: 10.1017/s0031182001001093. [DOI] [PubMed] [Google Scholar]
- 5.Tibayrenc M. Genetic subdivisions within Trypanosoma cruzi (Discrete Typing Units) and their relevance for molecular epidemiology and experimental evolution. Kinetoplastid Biol. Dis. 2003;2:12. doi: 10.1186/1475-9292-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brisse S, Verhoef J, Tibayrenc M. Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int. J. Parasitol. 2001;31:1218–1226. doi: 10.1016/s0020-7519(01)00238-7. [DOI] [PubMed] [Google Scholar]
- 7.Robello C, Gamarro F, Castanys S, Alvarez-Valin F. Evolutionary relationships in Trypanosoma cruzi: molecular phylogenetics supports the existence of a new major lineage of strains. Gene. 2000;246:331–338. doi: 10.1016/s0378-1119(00)00074-3. [DOI] [PubMed] [Google Scholar]
- 8.de Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Goncalves VF, Teixeira SMR, Chiari E, Junqueira ACV, Fernandes O, Macedo AM, et al. Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathog. 2006;2:e24. doi: 10.1371/journal.ppat.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macedo AM, Pena SD. Genetic variability of Trypanosoma cruzi: implications for the pathogenesis of chagas disease. Parasitol. Today. 1998;14:119–124. doi: 10.1016/s0169-4758(97)01179-4. [DOI] [PubMed] [Google Scholar]
- 10.Toledo MJ, de Lana M, Carneiro CM, Bahia MT, Machado-Coelho GLL, Veloso VM, Barnabé C, Tibayrenc M, Tafuri WL. Impact of Trypanosoma cruzi clonal evolution on its biological properties in mice. Exp. Parasitol. 2002;100:161–172. doi: 10.1016/s0014-4894(02)00003-6. [DOI] [PubMed] [Google Scholar]
- 11.Brisse S, Barnabé C, Bañuls AL, Sidibé I, Noël S, Tibayrenc M. A phylogenetic analysis of the Trypanosoma cruzi genome project CL Brener reference strain by multilocus enzyme electrophoresis and multiprimer random amplified polymorphic DNA fingerprinting. Mol. Biochem. Parasitol. 1998;92:253–263. doi: 10.1016/s0166-6851(98)00005-x. [DOI] [PubMed] [Google Scholar]
- 12.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran A.-N, Ghedin E, Worthey EA, Delcher AL, Blandin G, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 13.Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 14.Hartl DL, Clark AG. Principles of Population Genetics. 4th. Inc., Sunderland, MA: Sinauer Associates; 2007. [Google Scholar]
- 15.Agüero F, Zheng W, Weatherly DB, Mendes P, Kissinger JC. TcruziDB: an integrated, post-genomics community resource for Trypanosoma cruzi. Nucleic Acids Res. 2006;34:D428–D431. doi: 10.1093/nar/gkj108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agüero F, Al-Lazikani B, Aslett M, Berriman M, Buckner F, Campbell R, Carmona S, Carruthers I, Chan A, Chen F, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 2008 doi: 10.1038/nrd2684. Advanced Online Publication, October 17 2008; DOI:10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arner E, Kindlund E, Nilsson D, Farzana F, Ferella M, Tammi M, Andersson B. Database of Trypanosoma cruzi repeated genes: 20 000 additional gene variants. BMC Genomics. 2007;8:391. doi: 10.1186/1471-2164-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 20.The Comprehensive Perl Archive Network. 1995. http://cpan.org. Last accessed: October 27th, 2008.
- 21.Rockway J. Catalyst, Accelerating Perl Web Application Development. Birmingham, UK: Packt Publishing; 2007. [Google Scholar]
- 22.Agüero F, Verdún RE, Frasch AC, Sánchez DO. A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery. Genome Res. 2000;10:1996–2005. doi: 10.1101/gr.gr-1463r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stajich JE, Hahn MW. Disentangling the effects of demography and selection in human history. Mol. Biol. Evol. 2005;22:63–73. doi: 10.1093/molbev/msh252. [DOI] [PubMed] [Google Scholar]