Abstract
Transfer RNAs (tRNAs) represent the single largest, best-understood class of non-protein coding RNA genes found in all living organisms. By far, the major source of new tRNAs is computational identification of genes within newly sequenced genomes. To organize the rapidly growing collection and enable systematic analyses, we created the Genomic tRNA Database (GtRNAdb), currently including over 74 000 tRNA genes predicted from 740 species. The web resource provides overview statistics of tRNA genes within each analyzed genome, including information by isotype and genetic locus, easily downloadable primary sequences, graphical secondary structures and multiple sequence alignments. Direct links for each gene to UCSC eukaryotic and microbial genome browsers provide graphical display of tRNA genes in the context of all other local genetic information. The database can be searched by primary sequence similarity, tRNA characteristics or phylogenetic group. The database is publicly available at http://gtrnadb.ucsc.edu.
INTRODUCTION
Transfer RNA (tRNA) genes play an essential role in protein translation in all living cells. Among the numerous tRNA search programs created in the last 10 years, tRNAscan-SE (1) remains a popular standard for whole-genome annotation of tRNA genes. This PERL program uses the original tRNAscan program (2) and a linear sequence signal detection algorithm by Pavesi and colleagues (3) as pre-filters to obtain an initial list of tRNA candidates. The program then passes these candidates to a highly sensitive and selective covariance model search program (4) to obtain a final set of gene predictions that represent 99–100% of true tRNAs with a false positive rate of fewer than 1/15 gigabases (1).
To catalog the increasing number of predicted tRNA genes found in complete genomes, we developed the Genomic tRNA Database (GtRNAdb) as a repository for all identifications made by tRNAscan-SE. This database has been in regular use by the community for over 7 years, but never formally described. Recently, we updated the interface, content and search capabilities, justifying a new report of this improved resource. As before, the database provides summary statistics of predicted tRNA genes and the number of isotypes detected in each genome. Researchers can view tRNA genes by retrieving primary sequences, secondary structure information and isotype alignments. Alternatively, tRNA genes can now be viewed within the eukaryotic-specific UCSC Genome Browser (5) or similar microbial genome browsers (6). In addition, a new database search page and BLAST (7) server enable similarity studies of tRNA genes across species. To date, GtRNAdb contains 74 777 predicted tRNA genes derived from 36 eukaryotes, 55 archaea and 649 bacteria. Together with tRNAscan-SE, this public database provides an important information resource to the tRNA and genomics research communities.
DATABASE FEATURES
tRNA identification information
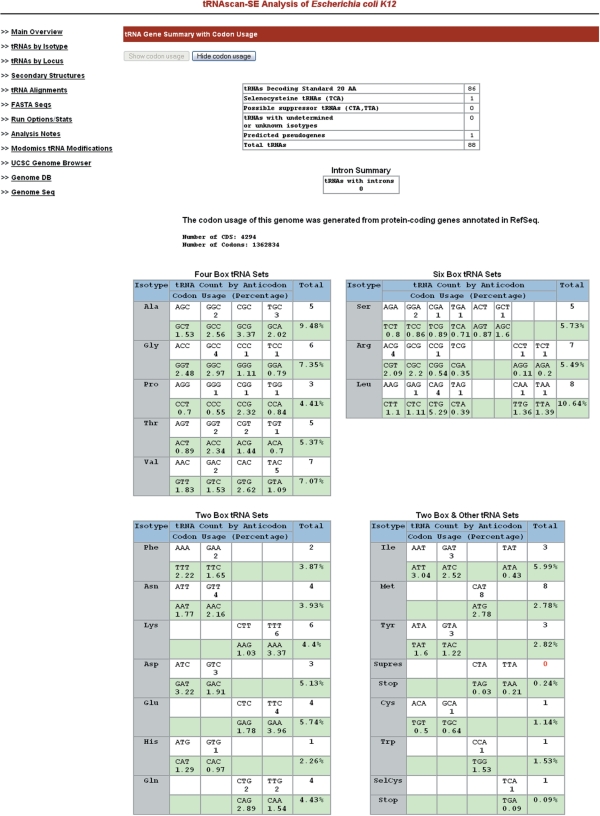
tRNAs from individual species can be selected from a full organism list on the GtRNAdb front page. Researchers can study the summary statistics of tRNA gene predictions from each genome, including the number of tRNAs with introns and the distribution of tRNAs belonging to each isotype. tRNA isotypes are grouped by ‘two-box’, ‘four-box’ or ‘six-box’ codon families, with highlighting colors to indicate potentially missing tRNAs (Figure 1). Users can study the frequency of tRNA genes in relationship to the codon usage, which is computed using protein gene annotations in NCBI RefSeq (8) for all prokaryotes and fungi, or obtained from the Codon Usage Database (9) for other eukaryotes. The GtRNAdb provides two viewing modes for gene lists: organized by isotype or by genome locus. Both views include tRNA gene and intron positions relative to the source chromosome (or plasmid); upstream and downstream sequence flanking the tRNA genes; and covariance model search scores that are broken down by contribution from primary sequence patterns versus secondary structures (this breakdown enables identification of some types of tRNA pseudogenes). If the eukaryotic or microbial genomes are available in external genome browsers (5,6), users can follow the provided links to study each tRNA within the context of neighboring genes. tRNA gene information can also be displayed and saved as plain text in the standard tRNAscan-SE output file format. In addition, researchers can download the tRNA sequences for each species in FASTA format, or as part of a full set for each phylogenetic domain.
Figure 1.
tRNA summary statistics with codon usage for Escherichia coli K12. Number of total tRNA genes and genes by isotypes and anticodons were provided by tRNAscan-SE (1) identification results. Protein-coding genes annotated in RefSeq (8) were used to compute codon usage of the genome. Side menus include links to detailed information for tRNA genes and external databases for gene analysis.
tRNA secondary structures and alignments
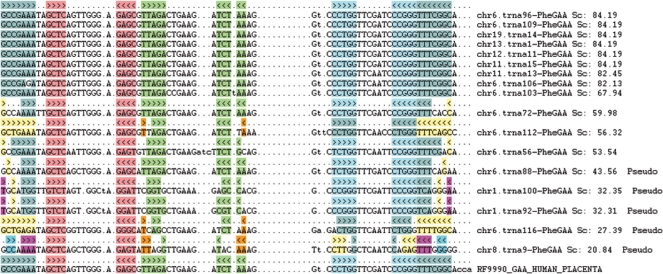
Although all mature non-organellar tRNAs form a general cloverleaf secondary structure, variations in the length of stem-loops exist. tRNAscan-SE (1) provides highly accurate secondary structure predictions via covariance model analysis (4) for each tRNA. These secondary structures can be viewed within GtRNAdb in linear string representations or as graphical two-dimensional images (Figure 2). To enable critical evaluation of lower-scoring tRNA identifications, the database also provides multiple sequence alignments across all tRNAs of the same isotype within a species. These structural alignments are constructed via alignment to domain-specific tRNA covariance models (4). Each stem-loop in the alignments is color-coded (similar to alignments found in Rfam 8.1) for easy viewing (Figure 3). For comparison to older reference tRNA sequences, multiple alignments also include aligned entries from the original Sprinzl tRNA database (10), when present from the same species and isotype.
Figure 2.
Secondary structure prediction of tRNA-GluCTC in chromosome III of Caenorhabditis elegans. (A) Linear string representation of secondary structure prediction generated within tRNAscan-SE by COVE (4). (B) Graphic representation of secondary structure prediction rendered by NAVIEW (14).
Figure 3.
Multiple sequence alignments of tRNA-PheGAA in Homo sapiens. Sequence alignments are grouped by identical secondary structures with the linear string representation listed on top of each block. Each color in the alignments codes for the base pairing of each stem loop in the secondary structure. The tRNA genes marked as ‘pseudo’ were identified as pseudogenes. The last tRNA RF9990_GAA_HUMAN_PLACENTA was retrieved from the Sprinzl tRNA database (10).
tRNA search and BLAST server
One of the goals in developing the GtRNAdb is to provide a tool for comparative analysis across multiple genomes. The search capabilities allow researchers to query the database with criteria including phylogenetic domain and clade, partial species name, chromosome or scaffold name, any combination of amino acids and anticodons, nucleotide identity at the −1 upstream position, number of introns and the existence of a genome-encoded terminal CCA sequence. Besides viewing results within the web browser interface, search results can be downloaded for further analysis, containing gene annotation and sequences. Researchers can use this search functionality to address various biological questions. For example, ‘which eukaryotes have predicted selenocysteine tRNAs in their genomes?’ By selecting the domain ‘Eukarya’ and amino acid ‘selenocysteine’, we find that there are 86 total selenocysteine tRNA predictions across 24 genomes such as human, mouse, horse, fruit fly and model legume Medicago truncatula.
Although genome-encoded anticodons starting with guanosine (G) or adenosine (A) are commonly used to decode codons ending with cytosine (C) or uridine (U), tRNAs with anticodons starting with A were not found in complete archaeal genomes (11). To search for possible exceptions, we selected the domain Archaea and all anticodons starting with A as the search criteria. The result shows that Ferroplasma acidarmanus includes a tRNA for leucine with anticodon AAG. Considering (i) the relatively low covariance model score of 45.65 bits as compared to the other tRNAs identified in the same genome and (ii) the absence of an expected leucine tRNA with anticodon GAG, this ‘flags’ either a potential sequencing error, or a target for further study in terms of post-transcriptional modification or RNA editing.
To search any given sequence directly against the tRNAs in the database, the tRNA BLAST server can be used. Options include searching for tRNA matches in all species, or only in one of the three domains of life. Standard BLAST options including expect value threshold and word size can be set for each query (7). Users can also enter advanced BLAST options in a free-text window. Pair-wise alignments are listed upon the completion of the search. If tRNA matches occur in genomes available in the external UCSC genome browsers (5,6), users can view tRNA hits within genomic context by clicking on the provided links.
Error and request tracking
In order to document tRNA gene predictions in a rapidly expanding list of completed genomes, most annotations in the database are automated without experimental verification or inspection against published literature. We acknowledge that there are exceptions to general anticodon-based isotype identification rules and other occasional errors due to post-transcriptional anticodon modification, unrecognized pseudogenes, some classes of short interspersed nuclear elements (SINEs) and other tRNA-derived sequences. In some cases, tRNA introns are also misidentified by automated searches (e.g. noncanonical introns found in many crenarchaeal species), which can cause incorrect determination of the anticodon and tRNA type. We have manually examined and corrected some of these errors (including crenarchaeal noncanonical introns and some tRNA-derived SINEs), yet we continue to search for new cases of obvious tRNA misidentification. We encourage feedback on any unaddressed discrepancies by submitting a report through our bug and request tracking system. We also welcome ideas for new features within the database, and often accept special requests for manually reviewed tRNA analyses from the user community. Users can monitor the progress of their requests and search through the development of other reports in the system.
FUTURE DIRECTIONS
Due to the design of a static web interface, the capability of data searching across genomes is currently limited. We plan to expand the database features by providing functionality to execute queries with more criteria such as ecotype of organisms, or allowing specification of sequence patterns at multiple positions within the tRNAs. Genes found via searches will be dynamically aligned with secondary structure information for comparative studies. Users will be able to download gene information in various file formats, including the BED format developed for the UCSC Genome Browser (5), and the Stockholm format used in Pfam (12) and Rfam (13) for multiple sequence and secondary structure alignments. We will also continue to update the database with new tRNA identifications as additional genomes are made available. Although the GtRNAdb generally focuses on collections of tRNAs from complete genomes, we encourage members of the research community to request analyses of draft or incomplete genomes.
FUNDING
Funding for open access charges: A gift from Hewlett-Packard via the UC Santa Cruz Center for Information Technology Research in the Interest of Society (CITRIS).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fichant GA, Burks C. Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol. 1991;220:659–671. doi: 10.1016/0022-2836(91)90108-i. [DOI] [PubMed] [Google Scholar]
- 3.Pavesi A, Conterio F, Bolchi A, Dieci G, Ottonello S. Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcriptional control regions. Nucleic Acids Res. 1994;22:1247–1256. doi: 10.1093/nar/22.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider KL, Pollard KS, Baertsch R, Pohl A, Lowe TM. The UCSC Archaeal Genome Browser. Nucleic Acids Res. 2006;34:D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grosjean H, Marck C, de Crecy-Lagard V. The various strategies of codon decoding in organisms of the three domains of life: evolutionary implications. Nucleic Acids Symp. Ser. (Oxf) 2007:15–16. doi: 10.1093/nass/nrm008. [DOI] [PubMed] [Google Scholar]
- 12.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruccoleri RE, Heinrich G. An improved algorithm for nucleic acid secondary structure display. Comp. Appl. Biosci. 1998;4:167–173. doi: 10.1093/bioinformatics/4.1.167. [DOI] [PubMed] [Google Scholar]