Abstract
OligoArrayDb is a comprehensive database containing pangenomic oligonucleotide microarray probe sets designed for most of the sequenced genomes that are not covered by commercial catalog arrays. The availability of probe sequences, associated with custom microarray fabrication services offered by many companies and cores presents the unequalled possibility to perform microarray experiments on most of the sequenced organisms. OligoArrayDb contains more than 2.8 probes per gene in average for more than 600 organisms, mostly archaea and bacteria strains available from public database. On average, 98% of the annotated genes have at least one probe which is predicted to be specific to its intended target in >94% of the cases. OligoArrayDb is weekly updated as new sequenced genomes become available. Probe sequences, in addition to a comprehensive set of annotations can be downloaded from this database. OligoArrayDb is publicly accessible online at http://berry.engin.umich.edu/oligoarraydb.
INTRODUCTION
Biology's entry into the genomic era during the past decade has led to new scientific challenges concerning not only the characterization of organism's genomes, but also their expression levels. Among the various types of investigations supported by DNA microarray technologies (1–7), transcriptome analyses are the most popular (8). One can monitor the expression levels of thousands of genes in parallel, and for eukaryotes, the expression level of splicing variants (9). The availability of flexible techniques for oligonucleotide synthesis in situ on microarrays (10–13) and commercial custom microarray fabrication service offers the unprecedented possibility to perform gene-expression studies on organisms not represented on catalog arrays from major manufacturers. This is the case for the vast majority of all sequenced bacteria.
The ensuing challenge for researchers is to determine the oligonucleotide probe sequences to be synthesized or spotted on the microarray. We and others have developed software to design probes for transcriptome analysis (14–21) but probe design can be a cumbersome task for inexperienced researchers. Probe design is the initial step in a microarray experiment and its quality will impact the final results thus many will prefer to use predesigned probe sets rather than taking the risk of doing a suboptimal design by themselves.
Several hundreds of organisms from archaea to eukaryotes have been sequenced so far and their genomes are available on public databases. If an organism proved to be of sufficient interest to be sequenced, then there is a chance that one may want to perform gene-expression studies on that particular organism. Thus, we have designed pangenomic oligonucleotide microarray probe sets for most of the sequenced genomes especially the ones not covered by commercial catalog arrays, and compiled them into a unique database.
Here, we present OligoArrayDb, a database containing oligonucleotide microarray probe sequences that allows, in conjunction with custom oligonucleotide microarray fabrication, to envisage any transcriptome analysis on sequenced organisms. The database is freely available online from http://berry.engin.umich.edu/oligoarraydb.
MICROARRAY PROBES DESIGN
Bacterial genomic sequences were obtained from the Genbank database (22). These sequences include plasmids when available. For eukaryotes, transcript sequences from chromosomes and organelle genomes were downloaded from Genbank (22), ENSEMBL (23) or TIGR-JCVI (24).
The probe design was done using the latest version of OligoArray (20) (v3.1, Rouillard, J.-M. unpublished data). Briefly, this program searches for specific probes at the genomic scale. The probe sequence is compared to all other expressed sequences from the same organism and the thermodynamic parameters (free energy and melting temperature) are computed for all possible hybridizations between the probe and perfect or nonperfect complementary sequences. If all of these values fall below a predetermined threshold, the probe is considered to be specific to its target. Probes are also selected to be unable to fold into stable secondary structures that may interfere with hybridization. Any probes with low sequence complexity or long stretches of the same base are rejected.
In terms of sensitivity, and specificity, the optimal size for an oligonucleotide grown directly on a microarray and used for gene-expression analysis is comprised between 50- and 60-mers (12,25). However, the 10 nt closest to the chip's surface seem to be not involved in hybridization due to steric interference (26); therefore, there is no reason to consider these nucleotides during the design process and specially during the specificity computation, as long as the corresponding sequence or any other kind of spacers with a sufficient length will be inserted between the target sequence and the chip surface during fabrication. According to these data, we have chosen to design probes with a size comprised between 45 and 47 nt. By using a range of length, the program can fit a narrower melting temperature (Tm) range in order to achieve better hybridization uniformity. The mean GC content was computed for each input sequence and the 5% extreme values on each side were filtered out. The remaining lower and higher values were selected to set the GC content range used during design. These values were also used to determine the optimal Tm. This approach allows us to design probes with consistent thermodynamic properties for all genes. Since hybridizations are usually carried out at or below 65°C, we use this temperature as a threshold to start considering non specific hybridization, but when a genome is highly GC rich, this value is slightly increased. In some cases, gene family members are so closely related that there is no way to discriminate between pairs of them. If no specific probe is found without cross-hybridization above the threshold, then all possible nonspecific hybridizations are reported in the output. Messenger RNAs from eukaryotes are polyadenylated and since this feature is used to anchor reverse transcription during probe labeling, we have limited the search space for probes to the last 1500 nt of the input sequences for eukaryotes. This limit is to prevent picking probes in a region that would eventually not be reverse transcribed in suboptimum experimental conditions. The input sequence is searched in a 3′ to 5′ direction to give preference to probe located as closely as possible to the messenger 3′-end. For archaea and bacteria where the mRNAs are not polyadenylated, the reverse transcription is usually primed with short random primers. This will lead to a better representation of the 5′-end of the mRNAs into the cDNA population. Thus, the input sequence is searched in a 5′ to 3′ direction to preferentially pick probes close to the messengers 5′-end.
Specific probes are ranked according to their position. For prokaryotes, the probe closest to the RNA 5′-end gets the highest rank, while for eukaryotes, the probe closest to the polyA tail gets the highest rank. If no specific probe exists for a given gene, then the probe with the lowest number of nonspecific targets is ranked first.
The specificity of the probes designed for this database was assessed as follow. Briefly, probes were designed for two different genomes, yeast and a bacteria ensuring that the probe specificity was computed against both genomes. Yeast total RNAs were labeled and hybridized to a microarray containing probes for both of these genomes. After hybridization, <0.3% of the bacterial probes (18 out of 6984) showed a signal above twice the background signal. Experimental details and results are reported on the OligoArrayDb homepage.
DATABASE CONTENT
In a first run, we have attempted to design up to three probes per transcript. In order to avoid any overlap between probes, we have chosen to impose a distance between probes at least equal to half the length of the mean probe length (23 nt). This implies that for relatively short sequences, it is not possible to design more than one or two probes. At the end, we have an average number of 2.82 probes per transcript successfully processed (n = 2 051 956 transcripts with probe(s) as of 1 September 2008). We have successfully designed at least one probe for >98% of all transcripts from all organisms processed (98.3%, n = 2 087 378 transcripts from 639 organisms). More than 94% of all transcripts with probe(s) have at least one specific probe (94.7%, n = 1 944 066 transcripts with specific probes). These percentages are 84% and 72% for transcripts with 2 and 3 specific probes, respectively. In the very few cases where the design failed, it is mostly due to input sequences shorter than the probe length or to monotonous sequences containing long stretches of the same nucleotide. Overall, OligoArrayDb contains 5 778 195 microarray probes representing 2 051 956 transcripts from 639 organisms or strains as of 1 September 2008. This database is regularly updated as new sequenced genomes are released.
AVAILABILITY
OligoArrayDb is publicly available with no restriction on its usage. It can be accessed online at http://berry.engin.umich.edu/oligoarraydb. For local implementation, data flat files and a building script (postgreSQL) are also available from the home page.
RETREIVING DATA FROM OLIGOARRAYDB
The home page gives a brief description of the database purpose, including the current counts on the number of genomes, transcripts and probes. More importantly, the home page lists the available genomes, separated in three columns according to their domain of origin, archaea, bacteria and eukaryote. Within each domain, strain or species are alphabetically sorted and linked to a probe set information page.
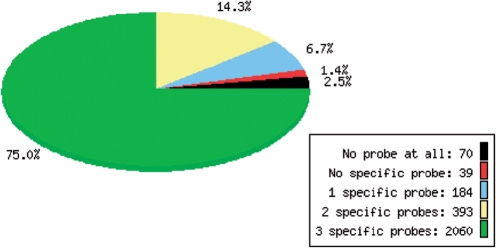
The probe set page gives details on the input sequence source, i.e. a link to the sequence file(s) used as input as well as relevant data on the sequence composition and number of transcripts. It also gives a link to the design parameters used to generate this particular probe set. Probe set composition is described in words and visually represented as a pie chart (see Figure 1 for a typical example). Lists of genes lacking probes or specific probes are also accessible from here. Finally, this page gives a link to the probe retrieval page.
Figure 1.
Number of transcripts with probes for Aeropyrum pernix K1. This genome contains 2746 genes. OligoArrayDB contains 2060 genes with three specific probes (75%, green), 393 genes with two specific probes (14.3%, yellow), 184 genes with one specific probe (6.7%, blue) and 39 genes with probes that are not specific (1.4%, red). The database does not contain any probes for 70 genes (2.5%, black).
The probe page offers the possibility to choose between retrieving the full data set or a customized one. One can choose between getting all the available probes (2.8 probes per gene on average; see above) or just one or two probes per gene. In this case, output probes will be selected according to their rank (see the probe design section above), highest rank first. Then one can choose the annotations to retrieve along with the probe sequence. Possible data are gene name, function, product and locus tag if available from the input sequence file. The probe size as well as its position on the input sequence is available. Position refers to the distance between the 5′-end of the probe and either 5′- or 3′-end (the later being mostly relevant to eukaryotes) of the target. One can choose also to report predicted thermodynamic data on hybridization of the probe to its intended target (free energy of hybridization, enthalpy, entropy and melting temperature of the hybrid). Finally, one can choose to report potential cross-hybridization targets if existing. From this column, users can tell whether a probe is specific to its target or not. The probe sequence comes with different options. One can choose between getting the probe alone (45- to 47-mer) or get it flanked by up to 15 nt either on the 5′- or 3′-end of the probe. The purpose of this sequence is to be used as a spacer to increase the distance between the surface and the sequence involved in the hybridization process. The user can choose either the real sequence contiguous to the probe sequence in the input sequence or a common artificial sequence like a polyT as recommend by Guo et al. (27). The data will be retrieved as a <TAB> delimited file ready to import into any text editing or spreadsheet software.
DISCUSSION
We have mainly focused our database on organism of medical, agronomical and industrial interest, leaving aside for now genomes well covered by commercial arrays, including the human genome. All organisms present in OligoArrayDb have been of enough interest to be covered by a sequencing project. But due to limited market, many of them will never be covered by commercial catalog microarrays. In situ synthesis technologies using digital photolithography (11,13) or inkjet printing (12) provide the ultimate flexibility in microarray fabrication as these processes rely only on probe sequence files. With OligoArrayDb, we provide probes for most of the organisms for which the genome sequence is known. The availability of a full set of probe sequences, associated with in situ synthesis offers now the unequalled possibility to perform custom microarray experiments on these organisms. The current database contains probe sets for more than 600 organisms as of 1 September 2008 and is weekly updated as new sequenced genomes are made available.
FUNDING
National Institute of Health (1 RO1 GM06854-01A1). Funding for open access charge: College of Engineering, University of Michigan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ehrenreich A. DNA microarray technology for the microbiologist: an overview. Appl. Microbiol. Biotechnol. 2006;73:255–273. doi: 10.1007/s00253-006-0584-2. [DOI] [PubMed] [Google Scholar]
- 2.Jares P. DNA microarray applications in functional genomics. Ultrastruct. Pathol. 2006;30:209–219. doi: 10.1080/01913120500521380. [DOI] [PubMed] [Google Scholar]
- 3.Kato H, Saito K, Kimura T. A perspective on DNA microarray technology in food and nutritional science. Curr. Opin. Clin. Nutr. Metab. Care. 2005;8:516–522. doi: 10.1097/01.mco.0000179166.33323.c3. [DOI] [PubMed] [Google Scholar]
- 4.Lettieri T. Recent applications of DNA microarray technology to toxicology and ecotoxicology. Environ. Health Perspect. 2006;114:4–9. doi: 10.1289/ehp.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmisano GL, Delfino L, Fiore M, Longo A, Ferrara GB. Single nucleotide polymorphisms detection based on DNA microarray technology: HLA as a model. Autoimmun. Rev. 2005;4:510–514. doi: 10.1016/j.autrev.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Walker MS, Hughes TA. Messenger RNA expression profiling using DNA microarray technology: diagnostic tool, scientific analysis or un-interpretable data? Int. J. Mol. Med. 2008;21:13–17. [PubMed] [Google Scholar]
- 7.Wiltgen M, Tilz GP. DNA microarray analysis: principles and clinical impact. Hematology. 2007;12:271–287. doi: 10.1080/10245330701283967. [DOI] [PubMed] [Google Scholar]
- 8.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 9.Bingham JL, Carrigan PE, Miller LJ, Srinivasan S. Extent and diversity of human alternative splicing established by complementary database annotation and microarray analysis. Omics. 2008;12:83–92. doi: 10.1089/omi.2007.0041. [DOI] [PubMed] [Google Scholar]
- 10.Beier M, Hoheisel JD. DNA microarray preparation by light-controlled in situ synthesis. Curr Protoc Nucleic Acid Chem. 2005;Chapter 12 doi: 10.1002/0471142700.nc1205s20. Unit 12.15. [DOI] [PubMed] [Google Scholar]
- 11.Gao X, LeProust E, Zhang H, Srivannavit O, Gulari E, Yu P, Nishiguchi C, Xiang Q, Zhou X. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 2001;29:4744–4750. doi: 10.1093/nar/29.22.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 13.Nuwaysir EF, Huang W, Albert TJ, Singh J, Nuwaysir K, Pitas A, Richmond T, Gorski T, Berg JP, Ballin J, et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen SH, Lo CZ, Tsai MC, Hsiung CA, Lin CY. The unique probe selector: a comprehensive web service for probe design and oligonucleotide arrays. BMC Bioinformatics. 2008;9(Suppl 1):S8. doi: 10.1186/1471-2105-9-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng S, Tillier ER. A fast and flexible approach to oligonucleotide probe design for genomes and gene families. Bioinformatics. 2007;23:1195–1202. doi: 10.1093/bioinformatics/btm114. [DOI] [PubMed] [Google Scholar]
- 16.Gasieniec L, Li CY, Sant P, Wong PW. Randomized probe selection algorithm for microarray design. J. Theor. Biol. 2007;248:512–521. doi: 10.1016/j.jtbi.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 17.He Z, Wu L, Li X, Fields MW, Zhou J. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 2005;71:3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Stormo GD. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics. 2001;17:1067–1076. doi: 10.1093/bioinformatics/17.11.1067. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Ying X. Mprobe 2.0: computer-aided probe design for oligonucleotide microarray. Appl. Bioinformatics. 2006;5:181–186. doi: 10.2165/00822942-200605030-00006. [DOI] [PubMed] [Google Scholar]
- 20.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouillard J-M, Herbert CJ, Zuker M. OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics. 2002;18:486–487. doi: 10.1093/bioinformatics/18.3.486. [DOI] [PubMed] [Google Scholar]
- 22.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shchepinov MS, Case-Green SC, Southern EM. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Z, Guilfoyle RA, Thiel AJ, Wang R, Smith LM. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]