Abstract
The Cell Line Data Base (CLDB) is a well-known reference information source on human and animal cell lines including information on more than 6000 cell lines. Main biological features are coded according to controlled vocabularies derived from international lists and taxonomies. HyperCLDB (http://bioinformatics.istge.it/hypercldb/) is a hypertext version of CLDB that improves data accessibility by also allowing information retrieval through web spiders. Access to HyperCLDB is provided through indexes of biological characteristics and navigation in the hypertext is granted by many internal links. HyperCLDB also includes links to external resources. Recently, an interest was raised for a reference nomenclature for cell lines and CLDB was seen as an authoritative system. Furthermore, to overcome the cell line misidentification problem, molecular authentication methods, such as fingerprinting, single-locus short tandem repeat (STR) profile and single nucleotide polymorphisms validation, were proposed. Since this data is distributed, a reference portal on authentication of human cell lines is needed. We present here the architecture and contents of CLDB, its recent enhancements and perspectives. We also present a new related database, the Cell Line Integrated Molecular Authentication (CLIMA) database (http://bioinformatics.istge.it/clima/), that allows to link authentication data to actual cell lines.
INTRODUCTION
Human and animal cell lines are widely used in basic and translational biomedical research, as they constitute a simple and representative model system for functional studies and identification of diagnostic tools and therapeutic targets. Each cell line has unique features and can be used for specific studies. It is therefore important to know characteristics of the cell lines and link the description to their availability. Presently, cell line information is mainly provided by collections on their sites. This has limitations, mainly related to the vast amount of collections and sites and to the heterogeneity of systems. Moreover, biological resources available from small collections or research laboratories can easily be missed.
For this reason, the Cell Line Data Base (CLDB) (1–3) was designed and implemented. Its development was funded by the Italian Ministry of University and Scientific and Technological Research (MURST) in the sphere of the initiatives for the strengthening of Italian infrastructures for biomedical research. It was developed according to a relational schema and populated by manual curation. It currently includes information on more than 6000 human and animal cell lines available from repositories and laboratories throughout Italy, as well as from major collections in other European countries. CLDB is not remotely accessible, but it can conveniently be searched via HyperCLDB (4), its hypertext version, that was later developed to allow for a user friendly access to the information. HyperCLDB is accessible online at the following URL: http://bioinformatics.istge.it/hypercldb/.
HyperCLDB consists of about 6000 pages describing cell lines and of more than 1000 index pages. These are based on terms included in the controlled vocabularies that are used to describe some of the main cell line features. All pages are built on a periodical basis by extracting information from CLDB by means of a set of purpose scripts. HyperCLDB is thus a static version of CLDB: pages are not created on-the-fly, but are consistently available as HTML files. This feature was conceived in order to allow archiving of the database contents in search engines repositories and it therefore significantly improved the likelihood of retrieving information. HyperCLDB allows for a trivial access to data based on main indexes and hypertext links connecting all pages. It includes all the contents of CLDB as well as links to external information sources, like OMIM, the Online Mendelian Inheritance in Man catalog (see http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim), and Medline, the US National Library of Medicine (NLM) database of biomedical bibliographic references (see http://www.ncbi.nlm.nih.gov/sites/entrez/). Since 1998, HyperCLDB has been used by biomedical researchers in order to retrieve accurate information on widely used human and animal cell lines, to identify distributors and to request cell lines from involved collections and laboratories. The success of this system is demonstrated by the number of links pointing to HyperCLDB from biomedical sites (e.g. some 600 hits are obtained when searching ‘hypercldb’ in Google) and by the high number of web site hits (in 2008, more than 20 000 unique visitors/month on average, see http://bioinformatics.istge.it/awstats/awstats.pl?config=www.biotech.ist.unige.it).
Recently, an interest was raised for a reference nomenclature for cell lines. Moreover, recent results, obtained using high-throughput genomic analyses, show that cross-contamination of human and animal cell lines is a repeated and frequent cause of scientific misrepresentation, and the assumption that the results obtained with the same cell lines by different researchers in different laboratories are fully comparable is often not true (5,6). Molecular methods for cell line authentication, such as fingerprinting, short tandem repeat (STR) profile and single nucleotide polymorphisms (SNPs) analysis, have been proposed in order to overcome this problem (7–11). In particular, an STR profile international reference standard for human cell lines was proposed by the leading cell banks from the United States, Europe, Asia and five large research institutes worldwide, who tested 253 human cell lines (12). Some well-known cell line banks, such as the American Type Culture Collection (ATCC, USA) and the Japanese Collection of Research Bioresources (JCRB, Japan) have made the results of STR profiling of the cell lines of their catalogs available (see, respectively, http://www.lgcpromochem-atcc.com/common/cultures/str.cfm and http://cellbank.nibio.go.jp/cellbank_e.html). A database including information on main kits for STR profiling and descriptions of related loci was also built (13). Now, the challenge is to build an integrated reference system able to link real cell lines maintained by different collections with authoritative molecular characterizations and to support cell culture authentication at a molecular level. A common reference portal is needed, where authentication data could be made available to the scientific community.
In this article, we present the architecture of CLDB, which was never published before, HyperCLDB, and perspectives of further developments. We also present a new database, the Cell Line Integrated Molecular Authentication (CLIMA) database, that allows to link available authentication data to actual cell lines described in CLDB.
MATERIALS AND METHODS
The CLDB was first created by using the Oracle database management system and it is now maintained by using mySQL public software. It resides on a Linux RedHat ES 4.1 server.
CLDB data structure
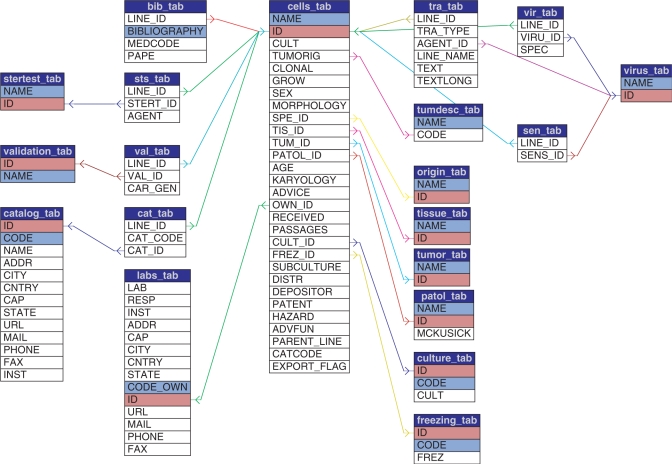
CLDB data structure was designed by taking into account the different structures of participating collections’ archives and databanks and with the goal of defining a standard for cell lines’ description. It includes terminologies taken from internationally recognized vocabularies. The database schema is shown in Figure 1. It includes the following types of tables:
A table (cells_tab) for cell lines features which are not encoded or, anyway, do not have a supporting vocabulary, such as culture type, morphology, karyology, tumorigenicity, clonality, growing conditions and all textual information, like advices, patents, hazard, distribution notes and depositor data. In this table, cell lines’ biological properties are also added, as free text.
Reference tables for controlled vocabularies on cell lines’ origin (including species and strains, tissues, tumors and pathologies), transformation (viruses and other transforming agents), culture and validation (culture and freezing media, sterility tests, validation assays) and ownerships and availability (laboratories, catalogs).
A few join tables that link cell lines to their encoded characteristics in a one-to-many relationship (e.g. one cell line can be validated by using different validation assays).
A table for bibliographic references. This is not a controlled vocabulary, but it is external to the cell lines table because of the one-to-many relationship between cell lines and papers.
Each table includes an auto-generated numeric identifier that is kept unchanged with the evolution of the database and that acts, once defined, as the unique identifier for that data (e.g. for that species, tissue, bibliographic reference, cell line, etc.). This feature is used in the HyperCLDB to generate URLs that are stable over time and do not change when the database is updated and the contents of single records is changed. As an example, let us consider the Drosophila melanogaster species. This is identified by the unique identifier 146 in CLDB species’ table. This identifier will never be changed: as a consequence, the list of CLDB cell lines derived from this species will always be included in a file named spe146.html. Analogously, cell lines associated with ‘amaurotic family idiocy, late infantile type/NCL late infantile’ (having McKucsick number 204500) will always be available in a file named pat270.html, since the numeric identifier for this pathology in the related table is 270. Finally, detailed information available in CLDB for the cell line 3T3 in the Interlab Cell Line Collection will always be available in a file named cl66.html because this cell line is inserted in the associated table with the unique identifier 66.
Figure 1.
CLDB database schema. Cells_tab includes features which are not encoded and textual information. Vocabularies are included in reference tables that are linked from cells_tab by their unique ids. Join tables are defined for linking catalogs and bibliographic references to cell lines since the same line can link to many catalogs and papers. The same apply to viruses table, since the same line can be transfected by more viruses.
It is important to point out that, as shown in the last example, the description of each cell line in CLDB is strictly linked to the collection/laboratory that provided the information. CLDB has distinct records even for those cell lines that, according to data providers, were taken from the same culture, but are conserved by different collectors. This is justified by the fact that contaminations and mutations can always occur and having the same origin is not a guarantee that the actual cell lines are the same. Similarly, sub-lines are described as cell lines derived by the original cell line, but distinct. Above cell lines’ unique identifier is thus only a feature of CLDB data structure. It links records in CLDB data tables referring to a specific cell line. As such, it does not describe the biological entity it refers to and it cannot document the evolution of the cell line. From a biological point of view, nor the name neither a numeric identifier could be seen as a ‘unique identifier’ of a cell line.
CLDB population
Cell lines’ data were gathered in many different formats through a tight collaboration with curators of collections and researchers of participating laboratories. It was then converted into the CLDB format and added to the database by using a purpose data curation application (not remotely available). The data curation application can be used by researchers with different aims and rights. The data manager is allowed to modify (insert, delete, change) all database contents. In particular, he can update, when needed, reference tables. Data curators can modify only those database contents that are related to their collection. A proper account is required to access management procedures. Insertion of a new cell line is carried out in two steps: first identification information is introduced, then additional data, such as sterility tests, validation assays and bibliography, can be added. Identification data can never be modified. Insertion of data is supported by some contextual help, e.g. pointing out mandatory fields and listing contents of controlled vocabularies. Data are double checked (automatically and manually) before its actual insertion into the database and online publication of data must be explicitly granted.
For consistency and interoperability aims, many data are coded according to controlled vocabularies. These are normally derived from internationally known reference codes and nomenclatures. These include, among the others, controlled vocabularies for pathologies, tumors, viruses and animal strains. Pathologies are coded according to the OMIM catalog of human genes and genetic disorders (14), tumors are defined according to the World Health Organization classification of tumors, and viruses are defined on the basis of the Universal Virus Database ICTVdB (http://www.ncbi.nlm.nih.gov/ICTVdb/) (15). References to animal strains are in accordance with current nomenclature, including the International Index of Laboratory Animals (16), the Mouse Genome Database (http://www.informatics.jax.org/) and the Rat Genome Database (http://ratmap.gen.gu.se/). Finally, bibliographic references are structured according to the US NLM specifications, and journals are defined on the basis of the list of serials indexed for online users.
HyperCLDB structure and generation
HyperCLDB consists in a set of HTML pages heavily interlinked. These files are stored in a unique directory. Their names are generated by using a standard procedure that takes into account the content type (i.e. cell line description, index of terms of a controlled vocabulary, index of cell lines annotated by a specific term of a vocabulary, etc.) and the contents unique ids (e.g. the cell line id, the term id, etc.). The same procedure is then used to add links between pages (e.g. from an index to a cell line description and vice versa), so that the hypertext is coherent. Examples of these names are: cl1000.html (description of the cell line with id 1000), spe27.html (index of cell lines derived from the species whose id is 27) and coll56.html (contact information for the collection having id 56). A series of specialized PHP scripts are available for building the different type of pages (e.g. one script creates detailed descriptions, one the pathology index, etc.). This allows to update the hypertext quickly, without re-creating it from scratch, when only a few changes are introduced in the database. HyperCLDB also includes links to molecular authentication data when available in the CLIMA database (see below). This link is only available in the description of cell line samples owned by the laboratory where authentications were performed.
CLIMA database
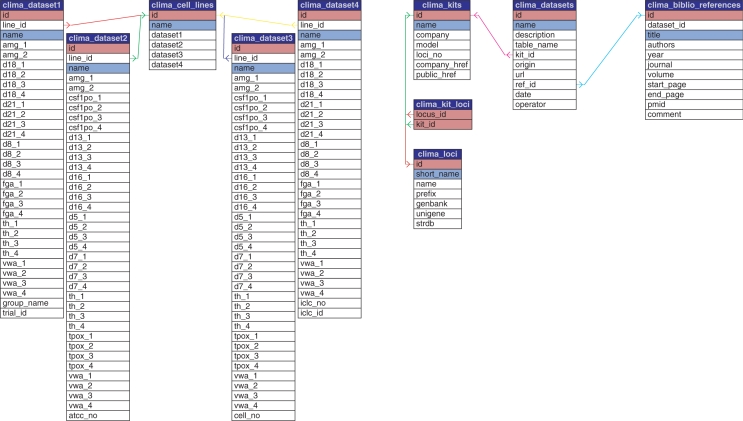
CLIMA was designed with the aim of representing a unique reference for validated molecular authentication of human cell lines, independently from the platforms (laboratory kits and/or sets of STR loci) used. For this reason, it includes STR profiling obtained by using different platforms and the end users can retrieve from the system data on cell line authentications performed by different cell banks. The schema of the database is shown in Figure 2. It includes data tables, where actual authentication data are stored, and metadata tables, where information on platforms and datasets are stored. Data tables consist in a general table, where information on all cell line names for which a molecular characterization exists are stored, and in one further table for each dataset including actual loci values. Each dataset may have a distinct set of loci and may include different additional information and thus has a special data structure. We decided to set up one distinct table for each dataset also because this choice simplifies data updates. Metadata tables include tables for the description of datasets, kits and related loci, and bibliographic information. This information is mainly used by applications for building query forms and carrying out searches.
Figure 2.
CLIMA database schema. Actual data tables (on the left) include the clima_cell_lines table, where information on cell line names for which a molecular characterization exist are stored, and one table for each dataset including STR profiles. Metadata tables (on the right) include tables for the description of datasets, kits and related loci, and bibliographic information.
RESULTS
CLDB contents
As of the end of July 2008, CLDB contains information on 6623 cell lines, 325 of which are not available for distribution. Among the remaining 6298 cell lines, 4918 are of human origin and 1380 of animal origin, from 80 different laboratories and collections. Among human cell lines, 990 are tumor derived (from 130 different tumors), and 1994 are developed from clinical specimens derived from patients affected with pathologies that, in our system, are defined, when appropriate, according to McKusick classification (299 different pathologies are described at present). By different transforming agents, 190 human cell lines are transformed by different agents, including viruses, oncogenes, chemical agents and radiations. Animal cell lines derived from 203 different species (when available, strain information is added and different strains are considered as different origins); 519 animal cell lines are of tumor origin (80 different tumors), while 537 cell lines are transformed by different transforming agents. This information was added to the system starting from 1992 and it was updated until 2003. Since then, the curation of the database was limited to the elimination of those cell lines that were no more available at the collection or laboratory. The only exception refers to the contents of the ICLC Cell Bank of our Institute, whose data are consistently added and regularly updated, based on the evolution of the cell line, i.e. passage number, and new findings on the cell line (i.e. new bibliographic references, further characterization based on newly available techniques, better classification of tumor).
HyperCLDB navigation
HyperCLDB permits the retrieval of detailed information on cell lines in various ways, including direct access to the description through the cell line name, free text search and navigation through the hypertext. In the latter case, starting points are represented by properties’ high-level indexes that include a list of all terms of related vocabularies. For example, the species high-level index includes a list of all species for which at least one cell line exists in the database. From these, users can access to low-level indexes that include lists of all cell lines having that particular property. For example, a species low-level index exists for any species listed in the high-level species index and it includes a list of all cell lines originated from that species. So, end users can choose any high-level index, click on one specific vocabulary term and reach the related low-level index, from which he can finally reach the description of the desired cell line.
At every stage of their navigation, end users can choose a different path, e.g. by moving from species navigation to tissue or pathology one. This behavior can easily be achieved because lists of cell lines available in low-level indexes include, beside their name, also some of their properties, namely those listed in other low-level indexes. Finally, from within the cell line description, users can move to all high- and low-level indexes that are somehow related to that cell line by using a similar method.
Each cell line description refers to a resource owned by a specific laboratory/collection. It consists of three parts: heading, general description, information on availability and handling. The heading includes name, species and strain (or ethnic group if human), tissue or organ of origin, tumor or pathology and transformation type, if any. Within the general description, the origin of the cell line is better defined, and further data are provided, given by the depositor and taken from literature; bibliographical references with links to PubMed are also included. Availability and handling data include information on how to obtain the cell line, culture and freezing media, handling of the cell line, quality control and specific characterization assays performed by the laboratory/collection, including STR profiling, extracted from the CLIMA database.
Opportune links to external information sources are an integral part of the hypertext. Links to OMIM permit the user to associate the cell lines derived from patients affected by genetic diseases to the corresponding OMIM detailed description. Links to Pubmed are also available, allowing researchers to retrieve information from reference papers, where cell lines were originally described, and from further papers of interest.
CLIMA contents
Presently, two platforms, that only share a limited number of loci, have been taken into account:
Commercial silica-gel-based purification kit (SGM) (Qiagen, Crawley, UK). The SGM kit comprises seven loci: human tyrosine hydroxylase (TH01, chromosome localization 11p15.5), human von Willebrand factor (vWF, 12p-12pter), D8S1179 (chromosome 8), D21S11 (21q11.2–21q21), human α Fibrinogen (FGA, 4q28) and D18S51 (18q21.3), human amelogenin (Xp22.1–22.3 and Yp11.2). For this platform, two datasets are available.
Commercial Promega PowerPlex® 1.2 system that is based on eight STR loci and the amelogenin gene. Loci are: D16S539 (16q24–qter), D7S820 (7q11.21–22), D13S317 (13q22–q31), D5S818 (5q23.3–32), human c-fms proto-oncogene for CSF-1 receptor gene (CSF1PO, 5q33.3–34), human thyroid peroxidase gene (TPOX, 2p24–2pter), human tyrosine hydroxylase gene (TH01, 11p15.5), human von Willebrand factor gene (vWF, 12p12–pter). For this platform, two datasets are available too.
CLIMA currently includes information on 1294 cell lines names. Four datasets are available, for 1737 distinct authentication assays. The following datasets are available:
Dataset 1: results obtained by leading cell banks and five large cancer research institutes by using platform 1 (12). This includes data on 223 cell lines.
Dataset 2: STR profiles made available by the ATCC and obtained by using platform 2. This dataset includes data on 670 cell lines and links to ATCC cell lines.
Dataset 3: STR profiles made available by the JBRC, using platform 2. This dataset includes data on 828 cell lines and links to JCRB cell lines.
Dataset 4: STR profiles obtained at our cell bank using platform 1. This includes data on 16 cell lines. This dataset also includes links to CLDB.
Metadata information is limited to the description of above datasets and platforms.
Access to CLIMA
CLIMA can be searched online at http://bioinformatics.istge.it/clima/ by using the purpose query forms that allow to search by name and by locus value. The search by name looks for all STR data that are assigned to cell lines whose name matches the submitted term. The search by name is case insensitive. Searching criteria include: exact match (the name must match the submitted string), truncation (the name must begin by the submitted string), soundex (name and string must match ‘by sound’: an algorithm largely applied in database systems is used) and punctuation removal (name and string are compared without taking into account punctuation, parentheses, spaces and a few extra characters). The search by locus looks in all available datasets for STR data corresponding to the searched locus values. Up to four values can be searched for each locus (with the exception of amelogenin that can only have two distinct values, one of which is always present) and combined by using logical AND and OR. Since end users can insert conditions on loci that are not available in all datasets, searches on each dataset are performed by using only those loci that are included in the dataset.
The search by name returns a summary of the query, the list of matching cell line names with indication of datasets including their STR profiles, and, for each dataset, a table including profiles and further information, like cell line catalog codes. Links to the original site from where the information was taken is connected to datasets names and links to the cell line description in the catalog is also provided, when available. The search by locus returns a summary of the search, a list of hits number for each dataset, and, again, a table for each dataset including STR profiles.
Although information included in the datasets can slightly differ, resulting tables have similar formats. The first column reports the cell line name and there is one further column for each locus in the dataset. Headings of columns referring to loci values include the name of the locus and a link to its description in STRBase (13).
Further developments
CLDB data management applications are under revision. When the new version will be released, collection curators will be allowed to manage their data remotely. Also, new automatic update procedures are under development: this will simplify and make more frequent data updates.
As to CLIMA, we intend to propose to collaborating cell line collections to work together to make it the cell line authentication database of a common European (and maybe international) portal for reference on authentication of established cell lines and lymphoblastoid cell lines.
We started an analysis of literature for identifying new STR profiles. We also are actively looking for authentication data provided by cell line collections. These activities will allow to include new datasets in the CLIMA database and to build a list of misidentified cell lines, including as well those for which only a suspect of misidentification exists. A warning tag will be added to the cell lines that are described in CLDB and have shown authentication ‘problems’. The tag will link to the list, where reference will be made to the publication(s) and/or websites describing the results of authentication. This will help researchers in checking whether the origin of the cell lines they are using should be confirmed.
Authentication of non-human cell lines, which are widely used in biomedical research, could also be performed by STR profiles from different species: the UgMicroSatdb (Unigene MicroSatellite database, a web-based, relational database of microsatellites present in unigene sequences covering 80 genomes) (17) will therefore be analyzed as a possible tool for further developments in this field.
CLIMA could become the cell line authentication database of a common European portal for reference on authentication of established cell lines. In this context, harmonization of information from various biology domains is essential. CLDB data structure was the result of a standardization effort among curators of involved collections. The CABRI (Common Access to Biological Resource and Information) EU project (see http://www.cabri.org/) took CLDB data structure into account for the definition of its minimum and recommended datasets for human and animal cell lines. We intend to move further along these lines: participation in new harmonization initiatives, such as Minimum Information for Biological and Biomedical Investigations (MIBBI) (18), is in our plans. A balance between the detailed datasets used by CLDB and the general information used by MIBBI projects must be found by abstraction.
Availability
Access to the HyperCLDB and the CLIMA is provided through the web site of the Bioinformatics group at the National Cancer Research Institute of Genoa. The main access points, respectively, are at the following URLs: http://bioinformatics.istge.it/hypercldb/, http://bioinformatics.istge.it/clima/.
Plans exist for the setting up of Web Services that would allow a programmable access to the data.
Currently, neither CLDB nor CLIMA complete datasets are made available to interested scientists for downloading. Special requests, justified on the basis of a collaboration proposal, should be addressed to the corresponding author.
FUNDING
Italian Ministry of Education and Scientific and Technological Research (MURST) [Interlab (1990–1992), LITBIO (2007–2010)]. Funding for open access charge: Italian Ministry of Education and Scientific and Technological Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Authors wish to thank all researchers and technicians who since 1992 contributed to the database either by inspiring its design, curating data or programming scripts. Special thanks go to Cesare Casirati, Beatrice Iannotta, Tiziana Ruzzon and Paola Visconti.
REFERENCES
- 1.Parodi B, Romano P, Aresu O, Manniello A, Vitiello E, Iannotta B, Ruzzon T, Santi L. The interlab project: data bases for biomedical research. Chem. Today. 1990;8:23–25. [Google Scholar]
- 2.Romano P, Aresu O, Iannotta B, Manniello A, Parodi B, Rondanina G, Ruzzon T. Interlab project databases: an effort towards the needs of a wider body of unskilled users. Binary. 1993;5:66–72. [Google Scholar]
- 3.Romano P, Manniello A, Campi G, Parodi B, Aresu O, Visconti P, Iannotta B, Rondanina G, Ruzzon T. Current status of the cell line data base. J. Exp. Clin. Cancer Res. 1995;14:6–7. [Google Scholar]
- 4.Manniello A, Ruzzon T. Cell line data base and HyperCLDB. Biotech. Knowl. Sources. 1996;9:3. [Google Scholar]
- 5.Freshney RI. Authentication of cell lines: ignore at your peril! Expert Rev. Anticancer Ther. 2008;8:311. doi: 10.1586/14737140.8.3.311. [DOI] [PubMed] [Google Scholar]
- 6.Lacroix M. Persistent use of ‘false’ cell lines. Int. J. Cancer. 2008;122:1–4. doi: 10.1002/ijc.23233. [DOI] [PubMed] [Google Scholar]
- 7.Dirks WG, Drexler HG. Authentication of cancer cell lines by DNA fingerprinting. Methods Mol. Med. 2004;88:43–55. doi: 10.1385/1-59259-406-9:43. [DOI] [PubMed] [Google Scholar]
- 8.Dirks WG, Faehnrich S, Estella IA, Drexler HG. Short tandem repeat DNA typing provides an international reference standard for authentication of human cell lines. ALTEX. 2005;22:103–109. [PubMed] [Google Scholar]
- 9.Dirks WG, Drexler HG. Authentication of scientific human cell lines: easy-to-use DNA fingerprinting. Methods Mol. Biol. 2005;290:35–50. doi: 10.1385/1-59259-838-2:035. [DOI] [PubMed] [Google Scholar]
- 10.Langdon SP. Characterization and authentication of cancer cell lines: an overview. Methods Mol. Med. 2004;88:33–42. doi: 10.1385/1-59259-406-9:33. [DOI] [PubMed] [Google Scholar]
- 11.Demichelis F, Greulich H, Macoska JA, Beroukhim R, Sellers WR, Garraway L, Rubin MA. SNP panel identification assay (SPIA): a genetic-based assay for the identification of cell lines. Nucleic Acids Res. 2008;36:2446–2456. doi: 10.1093/nar/gkn089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masters JR, Thomson JA, Daly-Burns B, Reid YA, Dirks WG, Packer P, Toji LH, Ohno T, Tanabe H, Arlett CF, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc. Natl Acad. Sci. USA. 2001;98:8012–8017. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruitberg CM, Reeder DJ, Butler JM. STRBase: a short tandem repeat DNA database for the human identity testing community. Nucleic Acids Res. 2001;29:320–322. doi: 10.1093/nar/29.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKusick VA. A Catalog of Human Genes and Genetic Disorders, 12th edn. Baltimore: Johns Hopkins University Press; 1998. Mendelian Inheritance in Man. [Google Scholar]
- 15.Büchen-Osmond C, Dallwitz MJ. Towards a universal virus database - progress in ICTVdB. Arch. Virol. 1996;141:392–399. doi: 10.1007/BF01718409. [DOI] [PubMed] [Google Scholar]
- 16.Festing M F W, editor. Available from: Centre for Mechanism of Human Toxicity. Leicester: 1993. International Index of Laboratory Animals 6th edn. [Google Scholar]
- 17.Aishwarya V, Sharma PC. UgMicroSatdb: database for mining microsatellites from unigenes. Nucleic Acids Res. 2008;36(Database issue):D53–D56. doi: 10.1093/nar/gkm811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor CF, Field D, Sansone S.-A, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz P.-A, Bogue M, Booth T, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat. Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]