Abstract
Small non-coding RNAs (sRNAs) carry out a variety of biological functions and affect protein synthesis and protein activities in prokaryotes. Recently, numerous sRNAs and their targets were identified in Escherichia coli and in other bacteria. It is crucial to have a comprehensive resource concerning the annotation of small non-coding RNAs in microbial genomes. This work presents an integrated database, namely sRNAMap, to collect the sRNA genes, the transcriptional regulators of sRNAs and the sRNA target genes by integrating a variety of biological databases and by surveying literature. In this resource, we collected 397 sRNAs, 62 regulators/sRNAs and 60 sRNAs/targets in 70 microbial genomes. Additionally, more valuable information of the sRNAs, such as the secondary structure of sRNAs, the expressed conditions of sRNAs, the expression profiles of sRNAs, the transcriptional start sites of sRNAs and the cross-links to other biological databases, are provided for further investigation. Besides, various textual and graphical interfaces were designed and implemented to facilitate the data access in sRNAMap. sRNAMap is available at http://sRNAMap.mbc.nctu.edu.tw/.
INTRODUCTION
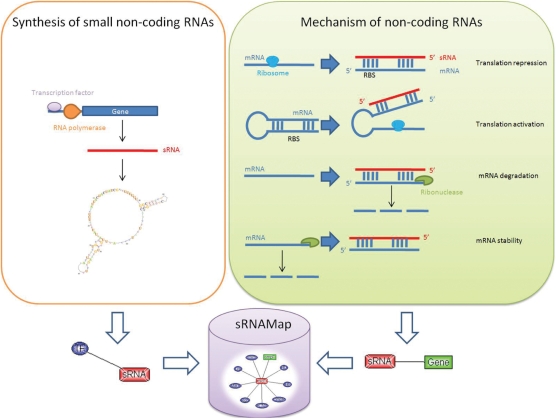
Small non-coding RNAs (sRNAs), which are discovered in many organisms ranging from bacteria to mammals, play important regulatory roles in cell physiology including regulation of cell development, cell death and chromosome silencing (1). Many of them regulate gene expression at a posttranscriptional level, either by acting as antisense RNAs, by binding to complementary sequences of target transcripts, or by interacting with proteins (2). Figure 1 depicts the synthesis and the functions of small non-coding RNAs. The transcription of sRNAs is regulated by transcription factors. Furthermore, sRNAs can play regulatory roles in translation repression, translation activation, mRNA degradation and mRNA stability.
Figure 1.
The synthesis and functions of small non-coding RNAs collected in sRNAMap.
EcoCyc (3) and RegulonDB (4) integrate biological knowledge of the transcriptional regulation in Escherichia coli, as well as knowledge on the organization of the genes and regulatory signals into operons in the chromosome. ASAP (5) is developed to store genome sequences in conjunction with associated annotations and functional characterization data. NONCODE (6) is an integrated knowledge database dedicated to non-coding RNAs. In addition, Storz et al. (7) used northern blotting analysis to document a total of 79 small RNAs in E. coli.
The increased investigations of important regulatory roles for sRNAs encoded far from their targets, acting on multiple targets, or both, has expanded interest in how to find such regulatory RNAs and how they work (8). Therefore, a resource collects the comprehensive annotation of small non-coding RNAs is crucial. We present an integrated database, sRNAMap, to collect the annotations of the sRNAs and the regulatory relationship between transcriptional regulator and sRNA, and between sRNA and its target genes. The design concept of the sRNAMap is illustrated in Figure 1. Additionally, more valuable information of sRNAs, such as the secondary structure of sRNAs, the expressed conditions of sRNAs, the expression profiles of sRNAs, the transcriptional start sites of sRNAs and the cross-links to other biological databases, are provided for further investigation. Besides, various textual and graphical interfaces were designed and implemented to facilitate the data access in sRNAMap.
DATABASE STATISTICS
The sRNAMap currently collects 397 sRNA genes, 62 regulator/sRNA regulations and 60 sRNA/target regulations in seventy microbial genomes. The detailed list of genome is given in Table S4. As given in Table 1, for instance, the number of experimentally validated sRNA genes in E. coli, Shigella boydii, Shigella flexneri and Yersinia pestis are 87, 35, 40 and 24, respectively. Table 2 gives the length distribution of the total known sRNA genes. Moreover, the sRNAMap analysed the transcriptional start sites of sRNA genes. Figure S3 (see Supplementary Materials) is the schematic diagram for the classification of transcription start sites of sRNA. In E. coli K-12 MG1655, 30 sRNAs have transcription start sites and 33 sRNAs have 49 putative transcription start sites, as given in Table S5.
Table 1.
The briefly statistics of small non-coding RNAs in sRNAMap
| Species names | No. of experimentally verified sRNAs |
|---|---|
| Escherichia coli | 87 |
| EnterobacteriaPhage VT2-Sakai genomic DNA | 1 |
| Enterobacter intermedius | 1 |
| Enterobacter cloacae | 1 |
| Enterobacter aerogenes | 1 |
| Yersinia pseudotuberculosis | 21 |
| Yersinia pestis | 24 |
| Yersinia mollaretii ATCC 43 969 | 1 |
| Yersinia intermedia ATCC 29 909 | 1 |
| Yersinia frederiksenii ATCC 33 641 | 1 |
| Yersinia enterocolitica | 23 |
| Yersinia bercovieri ATCC 43 970 | 1 |
| Shigella sonnei 046 | 38 |
| Shigella flexneri | 40 |
| Shigella dysenteriae | 33 |
| Shigella boydii | 35 |
| Serratia proteamaculans 568 | 2 |
| Salmonella typhimurium | 15 |
| Salmonella enteritidis | 2 |
| Salmonella typhi | 10 |
| Salmonella paratyphi A ATCC 9150 | 30 |
| Salmonella enterica Subsp. Arizonae | 1 |
| Salmonella choleraesuis SC-B67 | 4 |
| Photorhabdus luminescens TTO1 | 1 |
| Pectobacterium carotovorum | 2 |
| Pectobacterium atrosepticum SCRI1043 | 15 |
| Klebsiella pneumoniae | 4 |
| Klebsiella oxytoca | 2 |
Table 2.
Nucleotide length distribution of sRNA genes
| <100 | 100 ∼ 200 | 200 ∼ 300 | 300 ∼ 400 | 400 ∼ 500 | |
|---|---|---|---|---|---|
| No. of sRNAs | 334 | 436 | 68 | 58 | 12 |
DATA GENERATION
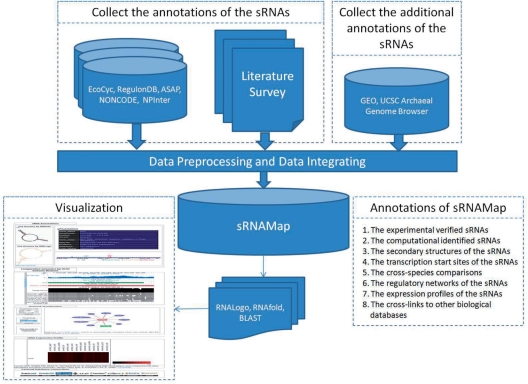
The data generation flow of sRNAMap database is depicted in Figure 2. The data generation flow comprises two major parts: (i) integration of external data sources and (ii) integration of annotated tools. We collect the sRNA information from a variety of biological databases, such as RegulonDB, ASAP and NONCODE. Information of sRNAs including the accessions, names, genomic location, species, descriptions and sequences were obtained. Furthermore, the regulator/sRNA regulations and sRNA/target regulations were obtained from RegulonDB and NPInter (9). In addition to collecting data from external databases, we gather the sRNA information by surveying literatures. Besides, RNA secondary structures, cross-species comparisons and 37 expression profiles of sRNAs were integrated into the database. 308 computationally identified sRNAs (10,11) and 114 computationally identified regulator/sRNA regulations and sRNA/target regulations (12,13) were obtained.
Figure 2.
The data generation flow of sRNAMap.
Gene Expression Omnibus (GEO) (14) is a database repository of high-throughput gene expression data and hybridization arrays, chips, microarrays. The expression profiles related to sRNA were obtained and integrated. Besides, UCSC Archaeal Genome Browser (15), which is a popular web-based tool for quickly displaying a requested portion of a genome at any scale, was integrated to provide the sequence conservation of sRNAs. RNAfold (16) was applied to fold the RNA secondary structures of sRNAs. Moreover, RNALogo (17), which presents a graphical representation of the patterns in an aligned RNA sequence family with a consensus structure, was integrated for presenting the sRNA families. The cross-links to other biological databases are provided for each sRNA in the database. The integrated external data sources, the linked external data sources and the integrated annotated tools are listed in Table S1, Table S2 and Table S3 (see Supplementary Materials), respectively.
INTERFACE
The sRNAMap provides a variety of interfaces and graphical visualization to present the plentiful information of sRNAs. Users can submit keywords or sequences to search the database. For each sRNA gene, the database provides the sequence, the genomic location, promoter information, secondary structures, literatures, annotations, expression profiles, sequence conservation and its transcriptional regulatory network. Additionally, the sRNAMap has the regulator/sRNA page and the sRNA/target page which provide the experimental conditions and the regulator/sRNA regulations and sRNA/target regulations. Figure S1 shows the interface of sRNA genes in sRNAMap.
sRNAMap also provides several browsing functions, such as the genome browser, the network browser, the expression profile browser, the computational sRNAs browser and the literature record browser (Figure S2, see Supplementary Materials).
DISCUSSIONS
sRNAMap is an integrated and comprehensive database comprising plentiful information about sRNA. Table 3 gives the comparison of sRNAMap with other databases related to sRNA including RegulonDB, ASAP, NONCODE, NPInter and Rfam (18). sRNAMAp aims on the annotation of small non-coding RNAs in microbial genomes, while Rfam mainly aims on the collection of non-coding RNA families and a variety of regulatory RNA structural motifs. Rfam currently collects 53 sRNA families. Our proposed sRNAMap collects 87 E. coli sRNAs and totally 397 sRNAs from 70 species. Moreover, sRNAMap also collects computational sRNA and supports information about RNA secondary structures, transcriptional start sites of sRNA and especially the expression profiles of sRNA. Consequently, we would like to say that sRNAMap provides more plentiful and effective information than Rfam and other databases in the aspect of sRNAs.
Table 3.
Comparing sRNAMap with other resources
| RegulonDB | ASAP | NONCODE | NPInter | Rfam | sRNAMap | |
|---|---|---|---|---|---|---|
| No. of sRNAs | 79 | 72 | 134 | 103 | 53 | 397 |
| No. of relations | ||||||
| Regulators/sRNAs | 16 | – | – | 50 | – | 62 |
| sRNAs/targets | 26 | – | – | 43 | – | 60 |
| No. of species supported | 1 (E. coli) | 59 (Microbial genomes) | 21 (Microbial genomes) | 1 (E. coli) | 248 | 70 (Microbial genomes) |
| Computational sRNAs supported | – | – | – | – | – | Yes |
| Secondary structure of sRNAs | – | – | – | – | Yes | Yes |
| Transcription start site of sRNAs | 1 typea | – | – | – | – | 5 typesa |
| Expression profiles supported | – | – | – | – | – | Yes |
| Transcriptional regulatory network | Regulators/sRNAs sRNAs/targets | – | – | – | – | Regulators/sRNAs sRNAs/targets |
| Sequence homology search | – | – | – | – | Yes | Yes |
aThe classification of transcriptional start sites of sRNA is described in Figure S1 (See Supplementary Materials).
AVAILABILITY
The sRNAMap database will be continuously maintained and updated. The database is now freely available at http://sRNAMap.mbc.nctu.edu.tw/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Council of the Republic of China (Contract No. NSC 96-3112-E-009-002, NSC 95-2311-B-009-004-MY3 and 97-2627-B-009-007); National Research Program for Genomic Medicine (NRPGM), Taiwan; MOE ATU (Partial). Funding for the open access publication charge: National Science Council of the Republic of China and MOE ATU.
Conflict of Interest statement: None declared.
ACKNOWLEDGEMENTS
Special thanks for financial support go to the National Research Program for Genomic Medicine (NRPGM), Taiwan.
REFERENCES
- 1.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 3.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasner JD, Liss P, Plunkett G.,, 3rd, Darling A, Prasad T, Rusch M, Byrnes A, Gilson M, Biehl B, Blattner FR, et al. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 2003;31:147–151. doi: 10.1093/nar/gkg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He S, Liu C, Skogerbo G, Zhao H, Wang J, Liu T, Bai B, Zhao Y, Chen R. NONCODE v2.0: decoding the non-coding. Nucleic Acids Res. 2008;36:D170–D172. doi: 10.1093/nar/gkm1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masse E, Majdalani N, Gottesman S. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 2003;6:120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 9.Wu T, Wang J, Liu C, Zhang Y, Shi B, Zhu X, Zhang Z, Skogerbo G, Chen L, Lu H, et al. NPInter: the noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res. 2006;34:D150–D152. doi: 10.1093/nar/gkj025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 11.Yachie N, Numata K, Saito R, Kanai A, Tomita M. Prediction of non-coding and antisense RNA genes in Escherichia coli with gapped Markov model. Gene. 2006;372:171–181. doi: 10.1016/j.gene.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 12.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider KL, Pollard KS, Baertsch R, Pohl A, Lowe TM. The UCSC archaeal genome browser. Nucleic Acids Res. 2006;34:D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang TH, Horng JT, Huang HD. RNALogo: a new approach to display structural RNA alignment. Nucleic Acids Res. 2008;36:W91–W96. doi: 10.1093/nar/gkn258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]