Abstract
ArrayExpress http://www.ebi.ac.uk/arrayexpress consists of three components: the ArrayExpress Repository—a public archive of functional genomics experiments and supporting data, the ArrayExpress Warehouse—a database of gene expression profiles and other bio-measurements and the ArrayExpress Atlas—a new summary database and meta-analytical tool of ranked gene expression across multiple experiments and different biological conditions. The Repository contains data from over 6000 experiments comprising approximately 200 000 assays, and the database doubles in size every 15 months. The majority of the data are array based, but other data types are included, most recently—ultra high-throughput sequencing transcriptomics and epigenetic data. The Warehouse and Atlas allow users to query for differentially expressed genes by gene names and properties, experimental conditions and sample properties, or a combination of both. In this update, we describe the ArrayExpress developments over the last two years.
INTRODUCTION
The ArrayExpress Repository is one of the three recommended international repositories to archive publication-related functional genomics data (1). The ArrayExpress Repository of array-based data was launched in 2002 (2), and currently it contains data from over 6000 experiments (studies) and approximately 200 000 assays. The ArrayExpress Warehouse of gene expression profiles was added in 2005 (3,4), and currently it includes data from over 600 studies and 20 000 samples. There have been three major developments in the last two years. First, the new ArrayExpress Atlas of Gene Expression provides experimental condition-based queries and an overview of gene expression across multiple experiments (Figure 1). Second, we have established a procedure to import data from the Gene Expression Omnibus (GEO) (5) and over 3000 GEO data series (GSE) have been imported and released via the ArrayExpress interface. Third, ArrayExpress has started to accept data from ultra high-throughput sequencing (UHTS) experiments and the first data sets have been made public. Other improvements include more flexible data access, web services, the support of the MAGE-TAB format (6), direct import from ArrayExpress into Bioconductor http://bioconductor.org/packages/2.3/bioc/html/ArrayExpress.html, a simpler submission process for large experiments, and use of ontologies to annotate and query the data.
Figure 1.
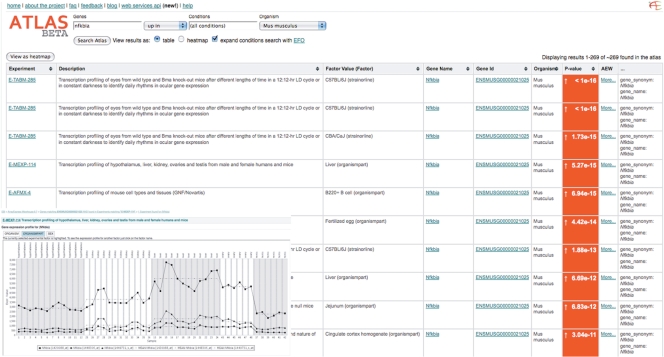
Table view from the ArrayExpress Atlas showing a query for up-regulation of the mouse gene Nfkbia in different experimental conditions. Columns from left to right: ArrayExpress experiment accession, experiment description, condition (factor value), gene name (linked to a gene view page), gene identifier, organism, P-value with an up/down indicator (red arrow for up, blue arrow for down) and a link to the ArrayExpress Warehouse for supporting data (inset bottom left).
ARRAYEXPRESS ATLAS OF GENE EXPRESSION
The ArrayExpress Atlas of Gene Expression (http://www.ebi.ac.uk/microarray-as/atlas/) allows the user to query for condition-specific gene expression across multiple data sets. The user can query for a gene or a set of genes by name, synonym, Ensembl identifier, GO term or, alternatively, for a biological sample property or condition, e.g. tissue type, disease name, developmental stage, compound name or identifier. Queries for both genes and conditions are also possible. For example, the user can query for all ‘DNA repair’ genes up-regulated in ‘cancer’. This returns a list of ‘experiment, condition, gene’ triplets each with a P-value and an up/down arrow characterizing the significance and direction of a gene's differential expression in a particular condition in an experiment. By default this list is ordered with most significant P-values on top. For example, a query for genes matching ‘nfkbia’, ‘up’ across all conditions in ‘Mus musculus’ returns a list of 230 matches in 30 different experiments and 100 different conditions where this gene is over-expressed (Figure 2). Each match is linked to detailed gene and experiment annotation in the Warehouse where expression profiles are presented as a graph of gene expression per sample and genes with similar expression profiles can be searched within an experiment. The raw experimental data files and complete experiment annotation are accessible through links to the ArrayExpress Repository.
Figure 2.
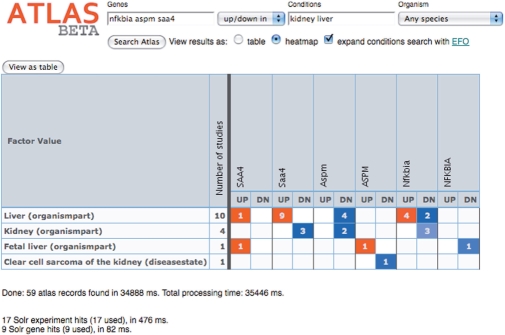
Heatmap view from the ArrayExpress Atlas showing an overview of expression of Nfkbia, Aspm and Saa4 genes in kidney and liver in all species.
As an alternative to presenting a ranked list of triplets, the Atlas heat map view provides a summarized table of gene expression patterns matching the query. Genes are listed in columns, conditions in rows, and the table cells are coloured red for over-expression, blue for under-expression and the number of independent studies where the pattern is observed is displayed within each cell (Figure 3). More complex queries combining several conditions are available via the advanced Atlas interface.
Figure 3.
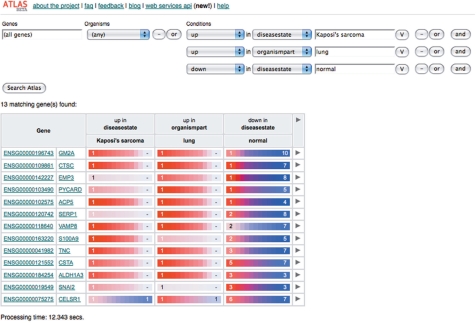
ArrayExpress Atlas advanced query interface showing all genes up-regulated in Kaposi's sarcoma diseased lung tissue and down-regulated in normal samples.
The Atlas data content is produced using a meta-analytical approach which uses the Bioconductor module limma (7). It is applied to each experiment in the ArrayExpress Warehouse to calculate moderated t statistics that simultaneously test the significance of differential expression in each condition versus the mean level of expression across all conditions for each gene. We then adjust the computed P-values to control the false discovery rate, using the Benjamini–Hochberg method (8), and aggregate the computed multiple test statistics for interpretation. The high quality of annotation and curation of experiments in the ArrayExpress Warehouse allows the application of this generic method to data from multiple platforms. Although the majority of the Atlas data at present is on the Affymetrix platform, the method is platform independent, and the Atlas also includes data from two-channel arrays. The ArrayExpress Warehouse and Atlas have a monthly data release with data derived from the ArrayExpress Repository content. Each release includes new experiments and a complete re-annotation of the genes present based on Ensembl and Uniprot, together with updated gene rankings.
DATA IMPORT FROM GEO
ArrayExpress now integrates gene expression data produced on Affymetrix and Agilent array platforms from GEO. This allows users to view and search GEO and ArrayExpress data from a common interface and access the data in the standard MAGE-TAB format. GEO data sets (GDS), using Affymetrix and Agilent catalogue arrays, are imported and converted from GEO SOFT to MAGE-TAB format. The data files are then checked and free text information is text mined using Whatizit (9) and a custom ArrayExpress dictionary derived from the NCI Thesaurus (10) with local extensions for non-human terms. Imported and text-mined datasets are then curated and the annotations mapped to ontology terms. GSE on Affymetrix and Agilent catalogue arrays are also imported and curated when GEO releases these as GDS or on request by a user community. The original GEO format accession numbers are retained as secondary accessions and links to the original data in GEO provided from the ArrayExpress interface. To date, there are 3700 GEO-derived experiments comprising more than 100 000 hybridizations present in the ArrayExpress repository. Code for parsing GEO SOFT format to MAGE-TAB, text-mining utilities and the custom dictionary are available (http://sourceforge.net/project/showfiles.php?group_id=120325). Mappings from SOFT to MAGE-TAB formats have been validated by GEO (T. Barrett, personal communication).
Once GDS are present in the ArrayExpress Repository, we apply Bioconductor quality metrics and perform scoring for MIAME compliance (11) to decide which data sets are appropriate for integration into the ArrayExpress Data Warehouse/Atlas. Curated GEO data that satisfy both the quality metrics and MIAME compliance and have curated experimental factors are included in the ArrayExpress Data Warehouse and consequently made available via Atlas queries.
NEW HIGH-THROUGHPUT SEQUENCING AND OTHER SUPPORTED DATA TYPES
ArrayExpress accepts data generated on all array-based technologies, including gene expression, protein array, ChIP-chip and genotyping. More recently, data from transcriptomic and related applications of UHTS technologies such as Illumina (SOLEXA Ltd, Saffron Walden, UK) (12), and 454 Life Sciences (Roche, Branford, Connecticut) are also accepted. For Solexa data FASTQ files, sample annotation and processed data files corresponding to transcription values per genomic location are submitted and curated to the emerging standard MINSEQE (http://www.mged.org/minseqe) and instrument-level data are stored in the European Short Read Archive (http://www.ebi.ac.uk/embl/Documentation/ENA-Reads.html). The first sequencing-based data sets to be made public are the transcriptomics landscape for Schizosaccharomyces pombe and epigenomics analysis of human (accession numbers E-MTAB-5 and E-TABM-482) (13,14). Spreadsheet templates are available for submission of these data from the ArrayExpress homepage.
The ArrayExpress Warehouse now includes gene expression profiles from in situ gene expression measurements, as well as other molecular measurement data from metabolomics and protein profiling technologies. Where in situ and array-based gene expression data are available for the same gene, these are displayed in the same view and links to the multispecies 4DXpress database of in situ gene expression (15) are provided.
OTHER DEVELOPMENTS
In addition to integrating processed data from multiple array platforms in the ArrayExpress Atlas, we have also performed a per platform integration using a re-annotation, data quality assessment and re-normalization approach. A large data set of more than 5000 hybridizations and 370 different biological conditions on the Affymetrix U133A platform is now available. The meta-analyses indicate that despite these data originating from multiple laboratories, the biological signal in these data is significantly stronger than the laboratory effects and new biological insights can be obtained from this approach (Lukk et al., manuscript in preparation). All raw and normalized data are available for this dataset (accession number E-TABM-185). Similar data sets have been produced for the mouse Affymetrix platforms U74Av2, MOE430A and 430 2.0 (accession numbers E-MTAB-26, E-MTAB-27, E-MTAB-28).
All ArrayExpress data are now available for download in MAGE-TAB format. To aid bioinformaticians and other users interested in large-scale functional genomics analysis, a Bioconductor package called ArrayExpress (http://www.bioconductor.org/packages/2.3/bioc/html/ArrayExpress.html) has been developed in collaboration with the Huber Group (EBI). This package allows the direct import of MAGE-TAB files into Bioconductor as native ExpressionSet objects, compatible with existing Bioconductor data analysis and visualization modules for this environment.
The ArrayExpress Atlas, Repository and Warehouse have web service APIs, enabling programmatic queries. ArrayExpress can also be queried along with all EBI core databases via the EBI general query interface ‘EB-eye’. The ArrayExpress submission tools, MIAMExpress and Tab2MAGE, are undergoing continuous improvement to facilitate submissions of large experiments, to work with MAGE-TAB files, and to accept UHTS-based transcriptomics data.
To improve queries of the ArrayExpress Atlas, we have developed an application ontology called the Experimental Factor Ontology (EFO). EFO version 0.6 (16) currently contains 1078 terms in an ‘is-a’ and ‘part-of’ hierarchy including diseases, multi-species anatomy, compounds and cell-type terms. It maps to several non-orthogonal ontologies, such as those for human anatomy, the Disease Ontology (17), the Cell Type Ontology (18) and the NCI Thesaurus (10). Use of the EFO allows tuning of the ontology based on analysis of user queries and provision of annotation at an appropriate level of granularity for the database content. The EFO is deployed in the Atlas interface where queries can be expanded via the hierarchies. For example, a query for the condition ‘cancer’ will also retrieve conditions ‘sarcoma’, ‘carcinoma’ and other cancers. The EFO is available from the EBI Ontology Lookup Server-OLS (19) and is available in OBO and OWL formats (http://www.ebi.ac.uk/microarray-srv/efo/).
FUTURE
There are two main challenges for ArrayExpress in the next two years—first to increase the volume of data in the Warehouse and Atlas, and second to build a standard pipeline for accepting and presenting UHTS-based data via the ArrayExpress Atlas. The underlying ArrayExpress architecture is undergoing a major re-development, which will provide improved performance to deal with large data sets and integration of multi-technology studies.
Currently, the ArrayExpress Warehouse/Atlas contains 10% of the data that are stored in the Repository and increasing this proportion in the next 12 months is a priority. An infrastructure is being developed that will allow the ArrayExpress curators to assess the appropriateness of new submissions for the Atlas at the point of the submission to the Repository, and to curate and load these data into a private Atlas instance. These data sets will then be automatically released in the Atlas as they become public. UHTS-based functional genomics data are already accepted and made available via the Repository. In the future, we will develop data analysis pipelines for these data sets for integration into the Atlas.
A new sample and experimental metadata database called the Bio Investigation Index (http://www.ebi.ac.uk/net-project/projects.html) is under development and will create a common structured representation and storage mechanism for metadata across several databases at the EBI. The new infrastructure also includes an emerging format for multi platform-based studies, ISA-TAB (20). Tools for creating ISA-TAB are under development, as well as converters between ISA-TAB and MAGE-TAB.
The ArrayExpress Atlas will undergo substantial user interface improvements, which will include a seamless integration of the Atlas with our online data analysis tool Expression Profiler (21). The code and ranking methodology will be made available via the Atlas documentation and a Bioconductor package. These will be the subject of a future publication. Finally, although the Repository, Warehouse and Atlas are distinct databases with different missions, a common user interface is under development and will be available in 2009.
FUNDING
The European Molecular Biology Laboratory; the European Commission grants FELICS (021902); EMERALD (LSHG-CT-2006-037689); DIAMONDS (LSHG-CT-2004-512143); BioSapiens (LSHG-CT-2003-503265); MolPAGE (LSHG-CT-2004-512066); MUGEN (LSHG-CT-2005-005203); Carcinogenomics (LSHB-2006-037712); Nutrigenomics (NoE-503630); Biotechnology and Biological Sciences Research Council (BB/E025080/1); the MAGE grant from National Human Genome Research Institute and National Institute of Biomedical Imaging and Bioengineering from the National Institute of Health (1 P41 HG003619-01). Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Wolfgang Huber and Audrey Kauffman for their work on the ArrayExpress and arrayQualityMetrics Bioconductor packages, Torsten Heinrich for assistance with import of in situ gene expression data and Tanya Barrett of GEO for assistance with SOFT to MAGE-TAB mapping. We would also like to thank the ArrayExpress Scientific Advisory Board.
REFERENCES
- 1.Ball CA, Brazma A, Causton H, Chervitz S, Edgar R, Hingamp P, Matese JC, Parkinson HE, Quackenbush J, Ringwald M, et al. Submission of microarray data to public repositories. PLoS Biol. 2004;2:1276–1277. doi: 10.1371/journal.pbio.0020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkinson H, Sarkans U, Shojatalab M, Abeygunawardena N, Contrino S, Coulson R, Farne A, Lara GG, Holloway E, Kapushesky M, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M, et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayner T, Rocca-Serra P, Spellman PT, Causton HC, Farne A, Holloway E, Liu J, Maier DS, Miller M, Petersen K, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. [Google Scholar]
- 8.Benjamini Y, Hochberg Y. Controlling for the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 9.Rebholz-Schuhmann D, Arregui M, Gaudan S, Kirsch H, Jimeno A. Text processing through web services: calling Whatizit. Bioinformatics. 2008;24:296–298. doi: 10.1093/bioinformatics/btm557. [DOI] [PubMed] [Google Scholar]
- 10.Fragoso G, de Coronado S, Haber M, Hartel F, Wright L. Overview and utilization of the NCI Thesaurus. Comp. Funct. Genomics. 2004;5:648–654. doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brazma A, Parkinson H. ArrayExpress service for reviewers/editors of DNA microarray papers. Nat. Biotechnol. 2006;24:1321–1322. doi: 10.1038/nbt1106-1321. [DOI] [PubMed] [Google Scholar]
- 12.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 13.Wilhelm B, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 14.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Gräf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haudry Y, Berube H, Letunic I, Weeber PD, Gagneur J, Girardot C, Kapushesky M, Arendt D, Bork P, Brazma A, et al. 4DXpress: a database for cross-species expression pattern comparisons. Nucleic Acids Res. 2008;36:D847–D853. doi: 10.1093/nar/gkm797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone J, Zheng-Bradley HJ, Rayner T, Parkinson H. Proceedings of the Eleventh Annual Bio-ontologies Meeting. San Diego, USA: Toronto; 2008. Developing an application focused experimental factor ontology: embracing the OBO Community; pp. 21–24. [Google Scholar]
- 17.Dyck P, Chisholm R. Proceedings of the Sixth Annual Bio-ontologies Meeting. San Diego, USA: Brisbane; 2003. Disease ontology: structuring medical billing codes for medical record mining and disease gene association; pp. 53–55. [Google Scholar]
- 18.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cote RG, Jones P, Apweiler R, Hermjakob H. The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics. 2006;7:97. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sansone SA, Rocca-Serra P, Brandizi M, Brazma A, Field D, Fostel J, Garrow AG, Gilbert J, Goodsaid F, Hardy N, et al. The first RSBI (ISA-TAB) workshop: “Can a simple format work for complex studies?”. OMICS. 2008;12:143–149. doi: 10.1089/omi.2008.0019. [DOI] [PubMed] [Google Scholar]
- 21.Kapushesky M, Kemmeren P, Culhane AC, Durinck S, Ihmels J, Körner C, Kull M, Torrente A, Sarkans U, Vilo J, et al. Expression Profiler: next generation—an online platform for analysis of microarray data. Nucleic Acids Res. 2004;32:W465–W470. doi: 10.1093/nar/gkh470. [DOI] [PMC free article] [PubMed] [Google Scholar]