Abstract
Circular permutation (CP) in a protein can be considered as if its sequence were circularized followed by a creation of termini at a new location. Since the first observation of CP in 1979, a substantial number of studies have concluded that circular permutants (CPs) usually retain native structures and functions, sometimes with increased stability or functional diversity. Although this interesting property has made CP useful in many protein engineering and folding researches, large-scale collections of CP-related information were not available until this study. Here we describe CPDB, the first CP DataBase. The organizational principle of CPDB is a hierarchical categorization in which pairs of circular permutants are grouped into CP clusters, which are further grouped into folds and in turn classes. Additions to CPDB include a useful set of tools and resources for the identification, characterization, comparison and visualization of CP. Besides, several viable CP site prediction methods are implemented and assessed in CPDB. This database can be useful in protein folding and evolution studies, the discovery of novel protein structural and functional relationships, and facilitating the production of new CPs with unique biotechnical or industrial interests. The CPDB database can be accessed at http://sarst.life.nthu.edu.tw/cpdb
INTRODUCTION
Circular permutation (CP) in the protein structure is a rearrangement of the amino acid sequence, such that the original amino- and carboxyl-termini of the polypeptide seem to be linked and new ones created elsewhere (1–4). This phenomenon was first observed in plant lectins 30 years ago (5). Since then, many natural cases have been discovered, including some carbohydrate-related enzymes and binding proteins, swaposins, transaldolases, FMN-binding proteins, glutathione synthetases, methyltransferases, ferredoxins, protease inhibitors, etc. (6). To reveal the effects of CP, many artificial circular permutants (CPs) have been generated, inclusive of the anthranilate isomerase, dihydrofolate reductase, T4 lysozyme, ribonucleases, aspartate transcarbamoylase, SH3 domain, ribosomal protein S6 and so on (7,8). The outcomes of these previous studies have indicated that CPs usually retain native structures and biological functions (3–5,9,10), although the stabilities and folding mechanisms might be altered (7,11,12). Since CP may sometimes increase the stability (13), activity or functional diversity (14–16) of proteins, it has been applied to trigger crystallization (13), improve enzyme activities (14), determine critical elements (17,18) and create novel fusion proteins (19–22).
In spite of these interesting properties and applications, there is still much uncertainty about the evolutionary mechanism, importance and natural prevalence of CP (7,9,23,24). Besides, even if there have been a few methods developed for the prediction of viable CPs, their performances were not well-assessed. The major cause of these uncertainties may be the lack of comprehensive resources of CP that can serve as a good base for studying it. This lack was basically because of the complicated rearrangement nature of circular permutation.
Conventional sequence and structural comparison methods employ collinear alignments and are inefficient to identify CP (9,25,26). To detect CP, several brilliant approaches have been developed, such as the sequence-based algorithms by Uliel et al. (27) and Weiner et al. (2), and the structure-based SHEBA (23), SAMO (26) and FASE (28). Sequence-based methods are fast, but they may miss many far-related CPs with low sequence similarities that can only be identified by structure-based methods (23), which are very time-consuming (6). We have developed an efficient CP-detecting procedure called CPSARST (Circular Permutation Search Aided by Ramachandran Sequential Transformation). The linear encoding methodology (29) and ‘double filter-and-refine’ strategy of CPSARST not only make it inherit the speed advantages of sequence-based methods but also retain the sensitivity to detect far-related CPs (6).
Here we present CPDB to be the first CP database. The primary data were screened from the Protein Data Bank (PDB) (30) by using CPSARST and then refined manually. There are currently 4169 nonredundant pairs of circular permutants recorded in the CPDB. CP pairs were grouped into CP clusters according to their direct and indirect CP relationships. Clusters were further grouped into folds and then classes based on their structural similarities. In addition, CPDB hosts a variety of tools and resources for studying CP, such as CP-based structural similarity search services, circularly permuted sequence/structure alignment and visualization tools, network representations of CP relationships, basic statistics of the properties of CPs and CP sites, and a well-organized list of CP-related literatures. Prediction methods for viable CPs described by Paszkiewicz et al. (31) are also implemented in the CPDB with some improvements. After an assessment, a measure known as ‘closeness’ (32) has been found successfully hitting 66.5% of the nonredundant CP sites in CPDB.
CP has long been used to study the folding mechanism of proteins. The evolutionary mechanism of CP itself is also interesting and has drawn many attentions (6). The information compiled in the CPDB is supposed to be helpful to move these research areas forward. Furthermore, most of the bioengineering and biotechnological applications of CP depend on a proper choice of position to create CP. The CP site information and viable CP site prediction methods provided by CPDB shall be advantageous to these fields.
CONTENTS AND METHODS
Identification of CP
Candidate pairs of circular permutants were first retrieved from a nonredundant PDB data set (26 349 polypeptides; see Supplementary List S1) by performing all-against-all searches with CPSARST (6) and then examined by visual inspections. After false cases were eliminated, the determined permutation sites of each pair were refined by the theoretically most accurate approach to identify CP (2,27), that is, generating all possible circularly permuted alignments to find the best way of aligning a pair of proteins. FAST (33) was applied as the structural alignment engine in this step. Finally, 4169 CP pairs consisting of 2238 proteins were identified. Among these cases, some bear multi-domain architectures with intact domain sequences, such as those reported in (34), but most of them are multi-domain proteins with one domain disrupted by CP or single-domain proteins.
There are two major categories of genetic mechanisms proposed to be responsible for CP (1). Duplication/deletion (9,35) and duplication-by-permutation models (1,36) both rely on independent events of gene duplication and partial deletion of terminal regions, while the latter one also emphasizes that an in-frame fusion had occurred along with the duplication. (2) Fusion/fission models (2,24,34) indicate that a pair of circular permutants were created by independent fusions of two smaller components, or, after a protein undergone fission, the resulting two distinct genes subsequently reassembled in a different order. Although it was reported by using sequence-based analyses that, for multi-domain proteins, fusion/fission mechanisms seem more dominant (34), whether this is also true for those permutations within single-domain proteins, however, remains uncertain. A large amount of new structural data has now been retrieved by CPSARST, including those of many functionally and/or structurally similar circular permutants with extremely low sequence identities. We hope that these data provided by CPDB can be helpful to elucidate more clearly the evolutionary mechanism of CP.
Categorization of circular permutants
Circular permutants in the CPDB were categorized in a hierarchical way. First, proteins with direct or indirect CP relationships were grouped into a ‘cluster’. For instance, if proteins A and B is a CP pair (designated as A↔B), B↔C is another CP pair and there is no significant CP relationship detected between proteins A and C, then A↔B and B↔C will be considered to have direct while A and C have indirect CP relationships. In this simple cluster (A↔B↔C), A and C may still be related by an unobvious CP, such as a very small permutation size, or they are just linear structural homologs. Next, structural similarities among representative proteins of each cluster, i.e. the most highly connected proteins, were calculated by FAST (33) and then a nearest-neighbor clustering algorithm (37) followed by manual adjustments were performed to group structurally similar clusters into the same ‘fold’. Finally, folds were classified into three classes, i.e. mainly-alpha, mainly-beta and alpha–beta mixed proteins according to their secondary structure elemental contents (Supplementary Data S2). The titles and descriptions of each level of categories were given based on the structural and functional information provided by the SCOP (38), PDB (30) and GO (39) databases.
Circularly permuted alignments and the visualization of CP relationships
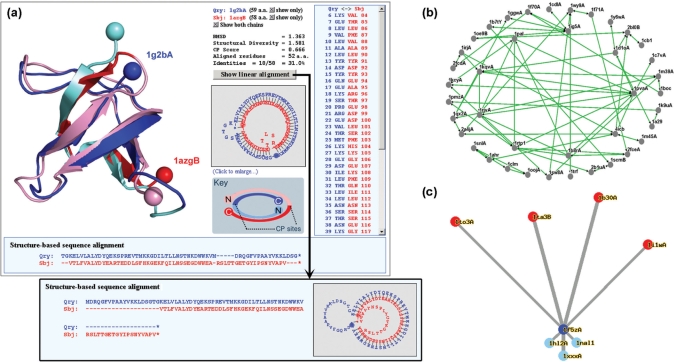
Circularly permuted structural alignments can be performed by FAST with suitable manipulations to the PDB file, as described in (6). We have implemented this strategy with a user-friendly way of visualization in the CPDB. As Figure 1a illustrates, the different locations of the termini and the position of CP sites can be easily recognized. The structure-based sequence alignment is shown in two different ways. The first is a plain text format in which unaligned regions are represented as gaps (-). The second is a graph with circularized text in which unaligned regions are represented as budding loops. Fewer loops or a smaller size of the loops stand for a larger number of residues that can be well aligned. If a pair of proteins is better aligned with a CP than without it, a CP relationship can be identified (2). If they can be well aligned both with and without a CP, they may be symmetric CPs (23). This circularized sequence alignment is especially helpful when the protein structures are too complicated for the user to trace their details.
Figure 1.
Various methods provided by CPDB for visualizing CP relationships among proteins. (a) Circularly permutated structure and sequence alignments. Cα atoms of terminal residues of the superimposed structures are shown as balls so that the different locations of termini, which are a property of CP, can be easily recognized. Two proteins are colored very differently. The boundaries between the lighter and darker colors are the positions of CP site. (b) Network view of a CP cluster. A CP cluster usually contains several CP pairs with direct or indirect linkages. Proteins with more complicated CP relationships are placed closer to the center of this network. (c) Star-like map of structural homologs. Query protein is at the center (the blue circle) with its circular permutants (red circles) radiating upwards and linear structural homologs (light blue circles) radiating downwards. Connecting lines are drawn in a way that their lengths are in proportion to the structural diversities (41) between proteins.
CPDB provides two methods to visualize the CP relationships among a group of proteins. For each CP cluster, a graphic ‘CP network’ was drawn by Osprey (40) (Figure 1b). For every protein, a star-like map was generated to show the structural diversities (41) from its circular permutants and linear homologs (Figure 1c).
Prediction of viable circular permutants
A measure known as residue closeness is useful for the identification of active site residues (32). Paszkiewicz et al. (31) have proven it also applicable to predict viable CP sites in protein structures and the accuracy is higher than that of relative side-chain area (RSA) or sequence conservation. We have re-implemented their methods of closeness and RSA. The results showed that 62.9% of the nonredundant CP sites in the CPDB could be successfully hit by using closeness and the successful rate of RSA is 60.4%. If we first added hydrogen atoms to PDB structures using the LEaP program of the Amber 6 package (42), the successful rate of closeness and RSA could be raised to 66.5 and 60.9%, respectively.
WEB INTERFACE
CPDB is implemented with MySQL 4 on a HP ProLiant ML570 machine with Linux operating system. A user-friendly web interface was developed by using PHP 5 scripting language, GD graphic library, JavaScript and Chime scripts for easy viewing and retrieval of the data. Figure 2 shows the navigation of the web pages:
Home page gives the background of CP and some basic statistics of the circular permutants recorded in CPDB.
Hierarchy browsing, batch browsing and the keyword search pages offer various methods for the users to obtain the information in which they are interested.
Protein page provides a variety of information including the functions, related references, protein and gene sequences, determined CP sites and CP site predictions. This page is cross-linked with many other pages of CPDB.
Alignment page offers novel visualization tools to examine circularly permuted sequences and structures.
CPSARST (6) and SARST (29) are provided to perform rapid structural similarity searches.
Literature list page offers greatly useful information about CP. Previous reports are well organized according to their purposes and methods. Both wet-lab experimental procedures and computational resources can be found through this page.
Figure 2.
Navigation of the CPDB. (a) Home page, (b) hierarchy browsing page, (c) search results page, (d) structural similarity search tools, (e) literature list page and (f) protein page. See Figure 1a for an example of the alignment pages.
FUTURE WORKS
Since the source of protein structures for the current release of CPDB is PDB, according to (6), the type of CP recorded in this database is basically the global CP (the unit of CP is the whole protein). However, partial CP (the CP is within a partial region of the protein) also exists in nature, even if some scientists consider it as ‘swap’ rather than CP (24). We have planned to enhance the ability of CPSARST to identify partial CPs by modifying its strategy and then update CPDB with the retrieved data. Once the information of partial CP is sufficient, a deeper understanding of the effects, importance and evolutionary mechanisms of CP shall be achievable. Besides, including these data will result in a larger training pool that is useful to develop more accurate predictors for viable circular permutants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council, Taiwan, R.O.C. [grant numbers 96-3112-B-007-006, 97-2752-B-007-003-PAE]. Funding for open access charge: National Science Council, Taiwan, R.O.C. [grant number 97-3112-B-007-007].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Margaret Dah-Tsyr Chang, Institute of Molecular and Cellular Biology, NTHU, for her insightful suggestions for the development of CPDB. We also thank Yu-Kwei Chang and Chun-Ting Yeh for their help in manually examining the raw data of CP pairs.
REFERENCES
- 1.Jeltsch A. Circular permutations in the molecular evolution of DNA methyltransferases. J. Mol. Evol. 1999;49:161–164. doi: 10.1007/pl00006529. [DOI] [PubMed] [Google Scholar]
- 2.Weiner J, III, Thomas G, Bornberg-Bauer E. Rapid motif-based prediction of circular permutations in multi-domain proteins. Bioinformatics. 2005;21:932–937. doi: 10.1093/bioinformatics/bti085. [DOI] [PubMed] [Google Scholar]
- 3.Tsai LC, Shyur LF, Lee SH, Lin SS, Yuan HS. Crystal structure of a natural circularly permuted jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes. J. Mol. Biol. 2003;330:607–620. doi: 10.1016/s0022-2836(03)00630-2. [DOI] [PubMed] [Google Scholar]
- 4.Ribeiro EA, Jr, Ramos CH. Circular permutation and deletion studies of myoglobin indicate that the correct position of its N-terminus is required for native stability and solubility but not for native-like heme binding and folding. Biochemistry. 2005;44:4699–4709. doi: 10.1021/bi047908c. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM. Favin versus concanavalin A: circularly permuted amino acid sequences. Proc. Natl Acad. Sci. USA. 1979;76:3218–3222. doi: 10.1073/pnas.76.7.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo WC, Lyu PC. CPSARST: an efficient circular permutation search tool applied to the detection of novel protein structural relationships. Genome Biol. 2008;9:R11. doi: 10.1186/gb-2008-9-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulaj G, Koehn RE, Goldenberg DP. Alteration of the disulfide-coupled folding pathway of BPTI by circular permutation. Protein Sci. 2004;13:1182–1196. doi: 10.1110/ps.03563704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinemann U, Hahn M. Circular permutations of protein sequence: not so rare? Trends Biochem. Sci. 1995;20:349–350. doi: 10.1016/s0968-0004(00)89073-8. [DOI] [PubMed] [Google Scholar]
- 9.Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr. Opin. Struct. Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 10.Vogel C, Morea V. Duplication, divergence and formation of novel protein topologies. Bioessays. 2006;28:973–978. doi: 10.1002/bies.20474. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Shakhnovich EI. Different circular permutations produced different folding nuclei in proteins: a computational study. J. Mol. Biol. 2001;306:121–132. doi: 10.1006/jmbi.2000.4375. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Wang J, Wang W. Transition states for folding of circular-permuted proteins. Proteins. 2004;57:153–171. doi: 10.1002/prot.20175. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz TU, Walczak R, Blobel G. Circular permutation as a tool to reduce surface entropy triggers crystallization of the signal recognition particle receptor beta subunit. Protein Sci. 2004;13:2814–2818. doi: 10.1110/ps.04917504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Z, Lutz S. Improving the catalytic activity of Candida antarctica lipase B by circular permutation. J. Am. Chem. Soc. 2005;127:13466–13467. doi: 10.1021/ja053932h. [DOI] [PubMed] [Google Scholar]
- 15.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 16.Todd AE, Orengo CA, Thornton JM. Plasticity of enzyme active sites. Trends Biochem Sci. 2002;27:419–426. doi: 10.1016/s0968-0004(02)02158-8. [DOI] [PubMed] [Google Scholar]
- 17.Anand B, Verma SK, Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 2006;34:2196–2205. doi: 10.1093/nar/gkl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebhard LG, Risso VA, Santos J, Ferreyra RG, Noguera ME, Ermacora MR. Mapping the distribution of conformational information throughout a protein sequence. J. Mol. Biol. 2006;358:280–288. doi: 10.1016/j.jmb.2006.01.095. [DOI] [PubMed] [Google Scholar]
- 19.Kojima M, Ayabe K, Ueda H. Importance of terminal residues on circularly permutated Escherichia coli alkaline phosphatase with high specific activity. J. Biosci. Bioeng. 2005;100:197–202. doi: 10.1263/jbb.100.197. [DOI] [PubMed] [Google Scholar]
- 20.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng. Des. Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- 21.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 22.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl Acad. Sci. USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung J, Lee B. Circularly permuted proteins in the protein structure database. Protein Sci. 2001;10:1881–1886. doi: 10.1110/ps.05801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uliel S, Fliess A, Unger R. Naturally occurring circular permutations in proteins. Protein Eng. 2001;14:533–542. doi: 10.1093/protein/14.8.533. [DOI] [PubMed] [Google Scholar]
- 25.Russell RB, Ponting CP. Protein fold irregularities that hinder sequence analysis. Curr. Opin. Struct. Biol. 1998;8:364–371. doi: 10.1016/s0959-440x(98)80071-7. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Wu LY, Wang Y, Zhang S, Zhang XS. Revealing divergent evolution, identifying circular permutations and detecting active-sites by protein structure comparison. BMC Struct. Biol. 2006;6:18. doi: 10.1186/1472-6807-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uliel S, Fliess A, Amir A, Unger R. A simple algorithm for detecting circular permutations in proteins. Bioinformatics. 1999;15:930–936. doi: 10.1093/bioinformatics/15.11.930. [DOI] [PubMed] [Google Scholar]
- 28.Vesterstrom J, Taylor WR. Flexible secondary structure based protein structure comparison applied to the detection of circular permutation. J. Comput. Biol. 2006;13:43–63. doi: 10.1089/cmb.2006.13.43. [DOI] [PubMed] [Google Scholar]
- 29.Lo WC, Huang PJ, Chang CH, Lyu PC. Protein structural similarity search by Ramachandran codes. BMC Bioinformatics. 2007;8:307. doi: 10.1186/1471-2105-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paszkiewicz KH, Sternberg MJ, Lappe M. Prediction of viable circular permutants using a graph theoretic approach. Bioinformatics. 2006;22:1353–1358. doi: 10.1093/bioinformatics/btl095. [DOI] [PubMed] [Google Scholar]
- 32.Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 2004;344:1135–1146. doi: 10.1016/j.jmb.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Weng Z. FAST: a novel protein structure alignment algorithm. Proteins. 2005;58:618–627. doi: 10.1002/prot.20331. [DOI] [PubMed] [Google Scholar]
- 34.Weiner J, III, Bornberg-Bauer E. Evolution of circular permutations in multidomain proteins. Mol. Biol. Evol. 2006;23:734–743. doi: 10.1093/molbev/msj091. [DOI] [PubMed] [Google Scholar]
- 35.Ponting CP, Russell RB. Swaposins: circular permutations within genes encoding saposin homologues. Trends Biochem Sci. 1995;20:179–180. doi: 10.1016/s0968-0004(00)89003-9. [DOI] [PubMed] [Google Scholar]
- 36.Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat. Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 37.Jain AK, Dubes RC. Algorithms for Clustering Data. New Jersey: Prentice Hall; 1988. [Google Scholar]
- 38.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 39.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu G. Top: a new method for protein structure comparisons and similarity searches. J. Appl. Cryst. 2000;33:176–183. [Google Scholar]
- 42.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]