Abstract
PRODORIC is a database that provides annotated information on the regulation of gene expression in prokaryotes. It integrates a large compilation of gene regulatory data including transcription factor binding sites, promoter structures and gene expression patterns. The whole dataset is manually curated and relies on published results extracted from the scientific literature. The current extended version of PRODORIC contains gene regulatory data for several new microorganisms. Major improvements were realized in the design of the web interface and the accessibility of the stored information. The database was further improved by the implementation of various new tools for the elucidation of gene regulatory interactions. Thus, the PRODORIC platform represents a framework for the interactive exploration, prediction and evaluation of gene regulatory networks in prokaryotes. PRODORIC is accessible at http://www.prodoric.de.
INTRODUCTION
In the last decade, the analysis and modeling of prokaryotic gene regulatory networks as basis of a systems biology approach to infection and biotechnological processes became of central interest (1,2). In this context network reconstruction requires reliable datasets of gene regulatory interactions, which are usually only available in the scientific literature. The fast accumulation of published gene regulatory data enhanced by the availability of numerous finished genomes and by high-throughput technologies fostered the development of structured repositories in the form of public databases.
Several specialized gene regulation databases with focus on one model organism or several organism groups were established (3–8). The PRODORIC database was released in 2003 as a universal data source covering gene regulation in prokaryotes with focus on pathogenic bacteria (9). In a manual curation process relevant data is extracted by constantly screening of the scientific literature. The main part of PRODORIC contains a unique collection of transcription factor binding sites (TFBSs) and their interacting transcription factors. Besides these regulatory interactions, promoter structures with transcriptional initiation sites and sigma factor binding sites were included. Moreover, gene expression data derived from published microarray experiments were integrated. An integral part of PRODORIC are aligned profiles of TFBSs for a certain regulator represented as positions weight matrices (PWMs) and sequence logos (10,11). Provided PWMs are useful tools for pattern matching, and thus for the prediction of unknown putative TFBSs in DNA sequences of interest. For this purpose PRODORIC is associated with the prediction tool Virtual Footprint that allows a PWM based scanning of sequences or even whole genomes for new regulator targets (12).
Here, we summarize the modifications and improvements of PRODORIC made in the recent years. This comprises a significant increase of data content and updates of our tools. Moreover, PRODORIC was further developed towards a database and bioinformatics tool platform combining data and software for the interactive browsing, prediction and evaluation of gene regulatory networks in prokaryotes.
DATABASE CURATION AND CONTENT
PRODORIC relies completely on published results with experimental validation and is not complemented with computational predicted data. The transformation of free-text data from the primary literature into structured information is constantly done manually by a team of curators. During the process of literature screening we observed that even refined PubMed searches with keywords like ‘gene regulation’ and ‘prokaryotes’ are not sensitive enough since important classification terms like ‘DNaseI footprint’ or ‘electromobility shift assay’ are not generally part of PubMed abstracts. Since these terms are often associated with figure captions we optimized the literature preselection and data mining tasks by use of the PDF search engine CaptionSearch (13).
The main content of PRODORIC was significantly increased to an overall number of nearly 3000 TFBSs. The number of promoter and operon structures as well as expression profiles increased concurrently (Table 1). The main portion of regulatory interactions is expectedly covered by the two model organisms Escherichia coli and Bacillus subtilis. Interestingly, these are followed by Pseudomonas aeruginosa and Staphylococcus aureus revealing the relevance of data from pathogenic bacteria. An other striking group of bacteria annotated recently are phototrophic bacteria like Rhodobacter sphaeroides and Synechococcus sp. DNA sequence elements like TFBSs or transcriptional start sites are usually mapped to fixed genomic positions. Consequently, PRODORIC is limitted to sequenced organisms with elucidated genome sequence. Therefore, finished genomes are imported from flat files into PRODORIC in a frequent process, so the number of available organisms has increased to 696 different bacterial genomes with a total of 1304 replicons.
Table 1.
Statistics of the PRODORIC content (september 2008)
| Organism | TFBSs | Genes | Regulons | PWMsa | Promoters | Profilesb |
|---|---|---|---|---|---|---|
| Escherichia coli | 1670 | 1045 | 90 | 88 (76) | 740 | 64 (4666) |
| Bacillus subtilis | 785 | 738 | 88 | 71 (53) | 493 | 34 (2488) |
| Pseudomonas aeruginosa | 197 | 264 | 33 | 22 (19) | 164 | 16 (1292) |
| Staphylococcus aureus | 106 | 39 | 9 | – | 29 | – |
| Rhodobacter sphaeroides | 38 | 33 | 4 | 3 (3) | 28 | – |
| Streptococcus pyogenes | 18 | 13 | 5 | 3 (2) | 11 | 19 (416) |
| Bradyrhizobium japonicum | 14 | 22 | 3 | 3 (3) | 11 | – |
| Synechococcus sp. | 13 | 6 | 2 | – | 13 | – |
| Rhizobium meliloti | 12 | 4 | 8 | – | 11 | 2 (198) |
| Others | 68 | 67 | 25 | 7 (7) | 86 | 13 (402) |
| Sum | 2921 | 2231 | 267 | 197 (163) | 1586 | 148 (9462) |
aThe non-redundant number of position weight matrices (number in parentheses).
bThe sum of genes that are linked to the profiles (number in parentheses).
For the purpose of pattern matching and prediction of potentially new transcription factor targets, a significant number of new PWMs were generated from aligned profiles of TFBSs (Figure 1). This PWM library provides the data basis for the PRODORIC associated prediction tool Virtual Footprint.
Figure 1.
Position weight matrix view of PRODORIC for the binding site of the Anr transcription faction from Pseudomonas aerugionosa.
DATABASE ACCESS
There are principally four different ways to access PRODORIC:
Submitting a database query via the supplied web forms.
Browsing through the content by the use of genome browser GBpro.
Exploring the regulatory network as visualized graph with the ProdoNet tool.
Accessing the database via webservices [Simple Object Access Protocol (SOAP) interface].
The previously developed PRODORIC web interface was significantly improved with regard to its design, handling and web browser support. Database queries with genes, proteins, TFBSs and PWMs can be submitted via web forms. We added new sections for searching promoters, expression profiles and whole regulons. Besides the regular web forms, various improved possibilities for interactive browsing of the database contents were implemented. The new version of the genome browser GBpro offers an improved presentation of gene regulatory features both as genome map and formatted sequence. In this context, the application of inline frames enabled a more convenient browsing through the database contents. We recently developed ProdoNet, a new visualization tool for the exploration of PRODORIC contents in an interactive graph view (14). This tool enables the detection and visualization of underlying gene regulatory networks to uncover the multiple levels of gene regulation like regulatory circuits and various network motifs. Moreover, ProdoNet allows for the mapping and visualization of sets of co-expressed genes to gene regulatory network graphs. A different method to query the database without using the webpages was implemented recently via the establishment of webservices using SOAP. These webservices enable a platform-independent access to PRODORIC and offer an interactive way for data integration which was realized first for the SYSTOMONAS and ROSY platforms (15,16). A more detailed description of the SOAP interface and application examples are available on the PRODORIC website.
PREDICTION OF GENE REGULATORY NETWORKS
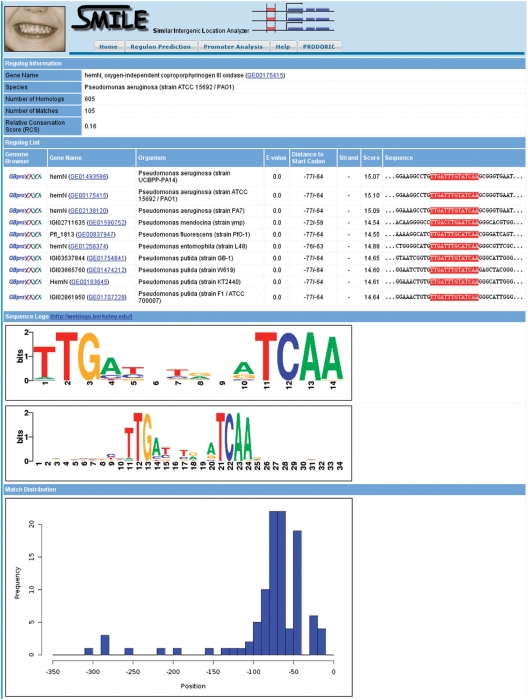
Although the PRODORIC core database excludes computationally predicted data, we follow the approach of a database assisted interactive prediction and validation of gene regulatory networks. Produced results are usually most accurate since they are based on the most recent set of data. For this purpose we developed Virtual Footprint, a tool for the prediction of potentially new transcription factor targets (12). Various search patterns can be defined by PWMs, IUPAC consensus strings or regular expressions. Complex bipartite patterns consisting of two subpatterns separated by a spacer are also possible. The integrated PWM library derived from the PRODORIC dataset was extended to 197 patterns corresponding to 163 different transcription factors (Table 1). The Virtual Footprint program allows the analysis of complete genomes with one PWM, which is called ‘regulon analysis’. In the other program mode ‘promoter analysis’, all available patterns are applied on one sequence. The new PRODORIC release 2009 was supplemented with a new tool called SMILE (similar intergenic location analyzer). Using this novel tool, the evolutionary conservation of Virtual Footprint matches can be further investigated by a comparative analysis of orthologous promoter sequences similar to a regulog analysis (17). In SMILE both sequence and positional conservation within an orthologous group of matches can be analyzed. This approach enables the evaluation of putative transcription factor targets and helps to rule out false-positive predictions (Figure 2).
Figure 2.
SMILE analysis using the Anr binding site in the promoter of the hemN gene. The results show both a high evolutional and positional conservation between the orthologous promoters (the list of matches was shortened).
CONCLUSIONS
PRODORIC is a manual curated data resource and bioinformatics tool platform about gene regulation and gene expression covering all sequenced prokaryotes. The whole system is supplemented with various browsing, prediction and validation tools representing a framework for the interactive analysis and visualization of gene regulatory networks. The manual curation process of PRODORIC will be continued. Mapping of gene regulatory interactions on sequenced genomes will be one of the most challenging task. The availability of reliable gene regulatoy networks will be essential for modeling approaches in systems biology.
FUNDING
The Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 578); German Bundesministerium für Bildung und Forschung (ERA-NET grant 0313936C). Funding for open access publication charge: Technical University of Braunschweig.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank our database curators Anne-Kareen Blechert, Tobias Knuuti and Roman Schubert for literature mining and trying to keep track with numerous published results. We are grateful to Karin and Lara Münch for assistance in web design. Finally, we would like to thank Bernd Hoppe for excellent technical assistance and financial management.
REFERENCES
- 1.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 2.Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, Garriga-Canut M, Serrano L. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gama-Castro S, Jiménez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Peñaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muñiz-Rascado L, Martínez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robison K, McGuire AM, Church GM. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- 5.Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumbach J. CoryneRegNet 4.0 – A reference database for corynebacterial gene regulatory networks. BMC Bioinformatics. 2007;8:429. doi: 10.1186/1471-2105-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazakov AE, Cipriano MJ, Novichkov PS, Minovitsky S, Vinogradov DV, Arkin A, Mironov AA, Gelfand MS, Dubchak I. RegTransBase – a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 2007;35:D407–D412. doi: 10.1093/nar/gkl865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Münch R, Hiller K, Barg H, Heldt D, Linz S, Wingender E, Jahn D. PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 2003;31:266–269. doi: 10.1093/nar/gkg037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'haeseleer P. What are DNA sequence motifs? Nat. Biotechnol. 2006;24:423–425. doi: 10.1038/nbt0406-423. [DOI] [PubMed] [Google Scholar]
- 11.Crooks GE, Hon G, Chandonia J, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 13.Mathiak B, Kupfer A, Münch R, Täubner C, Eckstein S. Improving Literature Preselection by Searching for Images. Lecture Notes in Computer Science. 2006;3886:18–28. [Google Scholar]
- 14.Klein J, Leupold S, Münch R, Pommerenke C, Johl T, Kärst U, Jänsch L, Jahn D, Retter I. ProdoNet: identification and visualization of prokaryotic gene regulatory and metabolic networks. Nucleic Acids Res. 2008;36:W460–W464. doi: 10.1093/nar/gkn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi C, Münch R, Leupold S, Klein J, Siegel I, Thielen B, Benkert B, Kucklick M, Schobert M, Barthelmes J, et al. SYSTOMONAS - an integrated database for systems biology analysis of Pseudomonas. Nucleic Acids Res. 2007;35:D533–D537. doi: 10.1093/nar/gkl823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pommerenke C, Gabriel I, Bunk B, Münch R, Haddad I, Tielen P, Wagner-Döbler I, Jahn D. ROSY – a flexible and universal database and bioinformatics tool platform for Roseobacter related species. In Silico Biol. 2008;8:177–186. [PubMed] [Google Scholar]
- 17.Alkema WBL, Lenhard B, Wasserman WW. Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 2004;14:1362–1373. doi: 10.1101/gr.2242604. [DOI] [PMC free article] [PubMed] [Google Scholar]