Abstract
We have constructed a database, AS-ALPS (alternative splicing-induced alteration of protein structure), which provides information that would be useful for analyzing the effects of alternative splicing (AS) on protein structure, interactions with other bio-molecules and protein interaction networks in human and mouse. Several AS events have been revealed to contribute to the diversification of protein structure, which results in diversification of interaction partners or affinities, which in turn contributes to regulation of bio-molecular networks. Most AS variants, however, are only known at the sequence level. It is important to determine the effects of AS on protein structure and interaction, and to provide candidates for experimental targets that are relevant to network regulation by AS. For this purpose, the three-dimensional (3D) structures of proteins are valuable sources of information; however, these have not been fully exploited in any other AS-related databases. AS-ALPS is the only AS-related database that describes the spatial relationships between protein regions altered by AS (‘AS regions’) and both the proteins’ hydrophobic cores and sites of inter-molecular interactions. This information makes it possible to infer whether protein structural stability and/or protein interaction are affected by each AS event. AS-ALPS can be freely accessed at http://as-alps.nagahama-i-bio.ac.jp and http://genomenetwork.nig.ac.jp/as-alps/.
INTRODUCTION
An increasing number of experiments have now demonstrated the biological significance of the diversification of protein structure generated by alternative splicing (AS), which results in diversification of interactions with other bio-molecules (1,2). For example, Dscam, a surface receptor required for both neuronal wiring and immune responses, is subjected to AS, producing thousands of isoforms. X-ray structural analyses of, and binding assays on, the amino-terminal four ecto-domains of two distinct Dscam isoforms have revealed that a sequence-variable region generated by mutually exclusive AS between the two isoforms is directly involved in isoform-specific homophilic binding (2). There are, however, huge numbers of AS variants that remain to be structurally and functionally characterized. To empower such experimental analyses, it is important to link information associated with protein structure, interaction and network to each AS variant, and also to predict the effect of each AS event (3). For such purposes, we have constructed a database, AS-ALPS (alternative splicing-induced alteration of protein structure).
There already exist a number of databases dealing with AS, many of which contain supplementary information regarding functional domains, such as those in InterPro (4). Although such information at domain-level resolution could be useful, it is often insufficient to infer functional change by AS, because amino acid sequences altered by AS (‘AS regions’) are often smaller than the typical domain size (3): most AS regions have a comparable size with that of secondary structure, super-secondary structure or module (5–7). Therefore, AS events need to be analyzed in terms of inter-residue and inter-atomic interactions between AS regions and other remaining regions of the protein. The three-dimensional (3D) structures of proteins are valuable sources of information for inferring the effects of AS on protein structure and interaction. Among AS-related databases, only AS-ALPS and ProSAS (8) integrate information about 3D protein structures. ProSAS, however, does not provide any information on protein interaction sites based on 3D structures, despite the importance of such information in detailed functional analysis of splice isoforms. This omission is probably because the main interest of ProSAS would be protein structure itself. Only AS-ALPS provides locations of interaction sites in the 3D structures of protein complexes with other proteins, nucleic acids or small ligand molecules, and also indicates whether AS regions include the residues that form such interaction sites. From this information, users can infer the effect of AS on protein interactions.
AS-ALPS also provides other unique and useful information. First, it provides the locations of hydrophobic cores, which are critical for protein structural stability, and can determine whether AS regions include residues forming these regions. From this information, users can infer the effect of AS on protein stability, which in turn helps to infer the effect of AS on protein interactions, i.e. to the extent that a stability change might alter such interactions. This information is not provided in any other AS-related databases, including ProSAS. Second, AS-ALPS provides links from AS-related transcripts to the protein network database KEGG (9). Users can then easily check which network and which node in the network can be influenced by AS. Third, AS-ALPS provides information regarding whether each AS variant mRNA can be subjected to nonsense-mediated mRNA decay (NMD), as predicted from the locations of premature termination codons (10). This information is only provided in one other AS-related database, H-DBAS (11). Taken together, these related data help users to select experimental targets from a huge collection of alternative transcripts, and to interpret the molecular mechanisms of AS-related network regulation.
DATABASE CONTENTS
AS regions
AS regions were detected as follows. First, a cluster of AS variant sequences was detected by aligning human (or mouse) full-length cDNA sequences from H-invDB (12) [or FANTOM (13)] and RefSeq (14) with the corresponding genomic DNA sequence stored at NCBI (Figure 1A). One of the AS variants, usually the most similar RefSeq entry to the counterpart in SwissProt (15), was defined as the reference transcript sequence. AS regions were then defined as regions of amino acid sequences that differ between the reference sequence and other AS variant sequences (Figure 1B). It should be noted that AS regions include not only amino acid sequence regions encoded by alternative exons but also those changed by frame shift due to AS. Appropriate comments were added to identify probably artifactual AS regions that were either derived from truncated terminal regions in incomplete transcripts or caused by inaccurate sequence alignment. The detected AS regions were also classified according to their sequence position and the type of change (insertion/deletion or substitution). In the case of substitution, we calculated the identity between the mutually replaced amino acid sequences, when they were comparable, allowing a measurement of the extent of the change.
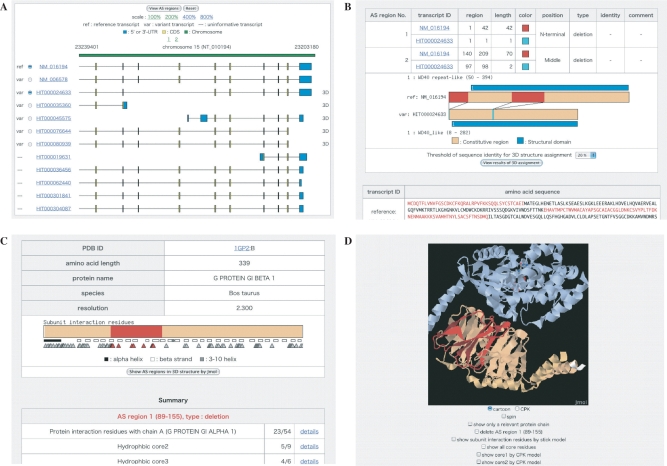
Figure 1.
Data display screens. (A) A cluster of AS variant sequences (CDS in yellow and UTR in blue) aligned with a chromosome sequence (green). A ‘3D’ mark in the rightmost column indicates that AS regions derived from the variant sequence in that row have 3D structure-related annotations. (B) Features of AS regions and their positional relationships to structural domains (blue). AS regions are colored by the type of change (deletion: red, insertion: cyan and substitution: violet). (C) Positional relationships of AS regions to hydrophobic cores and interaction sites detected in a relevant PDB entry. AS regions are shown in the same colors as in (B). Interaction sites are denoted with triangles. The positions of secondary structures are depicted with small open or filled boxes. (D) Spatial locations of AS regions in the 3D structure. A relevant protein chain with AS regions is shown in the same colors as in (C). Other interacting molecules are also shown in different colors.
Positional relationships of AS regions to structural domains
For each AS isoform, protein structural domains were assigned with the hidden Markov models in SUPERFAMILY (16). AS-ALPS provides positional relationships of AS regions to such structural domains (Figure 1B), from which users can determine whether each AS event changes a whole structural domain or just a part.
Positional relationships of AS regions to hydrophobic cores and interaction sites
The detailed procedures for annotating AS regions with 3D structural information were described in our previous paper (3). Briefly, a BLASTP homology search (17) against entries in PDB (18) was performed for each AS isoform. In the 3D structures for the hit PDB entries, hydrophobic cores and interaction sites were detected. Hydrophobic cores were defined as a spatial cluster of at least five amino acid residues meeting two conditions: (i) that solvent accessibility <0.2 and (ii) that >75% of surrounding atoms in contact with their side chain be carbon atoms (3). Interaction sites were defined as amino acid residues within 4 Å of other proteins, nucleic acids or ligands (19). AS-ALPS provides the positional relationships of AS regions to such hydrophobic cores and interaction sites (Figure 1C), from which users can infer the effects of AS on protein structure and interactions.
Possibility of NMD
AS-ALPS reports the possibility that each AS variant mRNA is subjected to NMD, as predicted from the locations of premature termination codons by the rule of Nagy and Maquat (10). It should be noted that some mRNAs have been experimentally demonstrated to be subjected to both NMD and translation (20). We have currently included even transcript sequences with a possibility of NMD in the analysis of AS regions.
ACCESS TO DATABASE
Search options
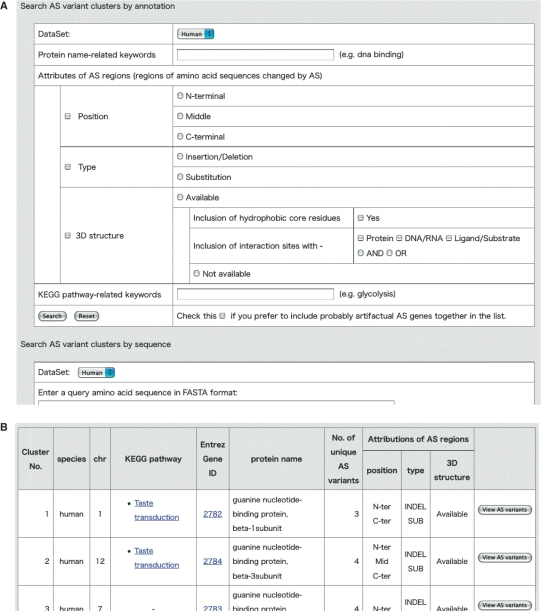
In the simple search form at the top of every page in AS-ALPS, either Transcript ID or Entrez Gene ID can be used as the search key. The advanced search page (Figure 2A) allows users to combine several search keys, such as species, protein name, attributes of AS regions (location, type and availability of 3D structure) and KEGG pathway name. If the 3D structure option is set to ‘available’, users can add search keys specifying whether AS regions include residues forming hydrophobic cores and what types of interaction sites the AS regions include. Search by amino acid sequence is also available, and is executed by the search engine FASTA (21). Users can also search by PDB ID. Search results are tabulated as in Figure 2B, where annotated information is briefly shown for each cluster of AS variants. The full list of genes subjected to AS can be browsed from the front page in the same format as that of the search result page, and also downloaded in tab-delimited format.
Figure 2.
The advanced search page (A) and an example of a table summarizing search results (B).
Data display screens
AS-ALPS has several data display screens for each type of information. First, clicking the ‘View AS variants’ button in the table of search results (Figure 2B) reveals the first screen, which shows a cluster of AS variants (Figure 1A). Next, after selecting one of variant transcripts, clicking the ‘View AS regions’ button leads to the second screen, which shows information regarding AS regions determined by comparing the amino acid sequences of the reference transcript and the user-selected variant (Figure 1B). AS regions are depicted in different colors depending on the type of change relative to the reference. Structural domains are also schematically represented with blue bars. In this example, one AS region of the deletion type (shown in red) is observed within the structural domain. Next, clicking the ‘View results of 3D assignment’ button leads to a list of assigned PDB entries accompanied by their amino acid sequence identities, as compared with the reference transcript. At that point, one of the PDB entries can be selected for viewing on the next screen, which shows the positional relationships of AS regions to hydrophobic cores and interaction sites detected in the selected PDB entry (Figure 1C). In this example, the AS region of the deletion type (shown in red) includes a part of the interaction sites (shown with triangles). Detailed information about interaction sites and hydrophobic cores is also provided at the bottom of the screen (Figure 1C). Next clicking the ‘Show AS regions in 3D structure’ button launches the Jmol 3D viewer (http://www.jmol.org/) (Figure 1D), showing the relevant protein chain in the same color as in Figure 1C, together with other interacting molecules. Spatial locations of hydrophobic cores can be viewed by using check boxes below the 3D viewer. The RasMol script for the 3D view can be downloaded. Branching off from the main route in AS-ALPS, the possibility of NMD can be checked in the page of detailed information on each transcript, which appears when the user clicks a transcript ID in the AS variant cluster page (Figure 1A).
Hyperlinks to external databases
AS-ALPS provides hyperlinks to several external databases, to allow easy access to related information. Each cluster of AS variants, which are assumed to be derived from the same gene, is linked to the relevant entry in Entrez Gene (22, 23) and to relevant protein networks in KEGG (9) (Figure 2B). On the linked KEGG pathway map, the query protein and other alternatively spliced proteins belonging to the same pathway are highlighted in yellow and pink, respectively. In the detailed information page of each transcript, transcript and genome sequence IDs are linked to the corresponding entries in their source databases. Ligands are linked to relevant entries in Het-PDB Navi (19).
CURRENT STATISTICS AND FUTURE DEVELOPMENTS
Detailed statistics of the source data and derived AS-ALPS data are reported on the statistics page accessed from the front page of AS-ALPS. Here, we present some statistics derived from the data set excluding probably artifactual AS regions. The current version of AS-ALPS, version 1.0, contains 11 926 genes subjected to AS (‘AS genes’) and 31 928 unique AS regions in human, and 7666 AS genes and 13 893 unique AS regions in mouse. The 3D structural information is available for 5067 (42.5%) and 2675 (34.9%) of AS genes in human and mouse, respectively, and is available for 10 056 (31.5%) and 3676 (26.5%) of unique AS regions in human and mouse, respectively. The fraction of AS genes with the 3D data in human (42.5%) has got much larger than that in our previous paper (about 7%) (3), although the procedure of analysis is not exactly the same as the previous one. Transcript data in AS-ALPS will be updated to fully recalculate the database contents at least annually if the source data have substantially changed. Structure data from PDB in AS-ALPS will be updated at least quaterly. To be able to follow the frequent updates of PDB, we are planning to automate AS-ALPS data updates. To enhance usability for researchers, we are also planning to add an option that allows user-uploaded transcript data to be included in the process of determining AS regions and annotating them with 3D structure information.
FUNDING
Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). Funding for open access charge: Genome Network Project from MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stetefeld J, Ruegg MA. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- 3.Yura K, Shionyu M, Hagino K, Hijikata A, Hirashima Y, Nakahara T, Eguchi T, Shinoda K, Yamaguchi A, Takahashi K, et al. Alternative splicing in human transcriptome: functional and structural influence on proteins. Gene. 2006;380:63–71. doi: 10.1016/j.gene.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Go M. Correlation of DNA exonic regions with protein structural units in haemoglobin. Nature. 1981;291:90–92. doi: 10.1038/291090a0. [DOI] [PubMed] [Google Scholar]
- 6.Noguti T, Sakakibara H, Go M. Localization of hydrogen-bonds within modules in barnase. Proteins. 1993;16:357–363. doi: 10.1002/prot.340160405. [DOI] [PubMed] [Google Scholar]
- 7.Shinoda K, Takahashi K, Go M. Retention of local conformational compactness in unfolding of barnase: contribution of end-to-end interactions within quasi-modules. Biophysics. 2007;3:1–12. doi: 10.2142/biophysics.3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birzele F, Kuffner R, Meier F, Oefinger F, Potthast C, Zimmer R. ProSAS: a database for analyzing alternative splicing in the context of protein structures. Nucleic Acids Res. 2008;36:D63–D68. doi: 10.1093/nar/gkm793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 11.Takeda J, Suzuki Y, Nakao M, Kuroda T, Sugano S, Gojobori T, Imanishi T. H-DBAS: alternative splicing database of completely sequenced and manually annotated full-length cDNAs based on H-Invitational. Nucleic Acids Res. 2007;35:D104–D109. doi: 10.1093/nar/gkl854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imanishi T, Itoh T, Suzuki Y, O’Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi A, Iida K, Matsui N, Tomoda S, Yura K, Go M. Het-PDB Navi.: a database for protein-small molecule interactions. J. Biochem. 2004;135:79–84. doi: 10.1093/jb/mvh009. [DOI] [PubMed] [Google Scholar]
- 20.Dreumont N, Maresca A, Boisclair-Lachance JF, Bergeron A, Tanguay RM. A minor alternative transcript of the fumarylacetoacetate hydrolase gene produces a protein despite being likely subjected to nonsense-mediated mRNA decay. BMC Mol. Biol. 2005;6:1. doi: 10.1186/1471-2199-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]