Abstract
The Human Genome Variation database of Genotype to Phenotype information (HGVbaseG2P) is a new central database for summary-level findings produced by human genetic association studies, both large and small. Such a database is needed so that researchers have an easy way to access all the available association study data relevant to their genes, genome regions or diseases of interest. Such a depository will allow true positive signals to be more readily distinguished from false positives (type I error) that fail to consistently replicate. In this paper we describe how HGVbaseG2P has been constructed, and how its data are gathered and organized. We present a range of user-friendly but powerful website tools for searching, browsing and visualizing G2P study findings. HGVbaseG2P is available at http://www.hgvbaseg2p.org.
INTRODUCTION
Genetic association studies provide a means to explore the genetic basis of complex traits, such as disease and drug response. Recent improvements in genotyping technologies and in sample biobanking have dramatically increased the scale and the accuracy of the data being produced. The reporting of results from such studies is, however, far from optimal; they are typically disseminated in diverse and disconnected databases, journals and meetings. Negative studies are all too often not reported at all (1). Consequently, there is no convenient way to gather together, compare and contrast findings from comprehensive subsets of related studies. This presents a major problem for the field, since association studies produce both positive and negative signals that may be real or false, and which can only be resolved by comparing independently generated data sets. The situation is further compounded by the recent emergence of large genome-wide association studies (GWAS) data sets, which involve the study of many thousands of subjects. For example, in the past year alone, NHGRI's GWA catalogue (http://www.genome.gov/26525384) lists over 100 studies for dozens of diseases and traits. These enormous studies bring extra complications with respect to data handling, data sensitivity and statistical interpretation.
The association study field thus requires new and effective means for helping researchers locate and compare existing data relevant to their disease or gene of interest, in an environment wherein negative studies can be reported as easily and quickly as positive studies. Public databases of genetic association data provide an obvious potential solution to this problem, especially if data deposition were encouraged by research-funding agencies and scientific journals. Two archival databases of this type do exist, namely dbGaP (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) (2) and EGA (http://www.ebi.ac.uk/ega/), as do a few small disease-specific initiatives (3–5). However, none of these bring together a globally comprehensive list of GWAS studies in a platform designed to also receive direct submission of any number of smaller studies. To provide the field with this missing capability, we recently released the Human Genome Variation Genotype to Phenotype database (HGVbaseG2P).
HGVbaseG2P represents the world's first central database for summary-level (i.e. not person-specific) genetic association data concerning any and all traits. It has been populated with an initial series of GWAS data sets acquired from other public depositories, and we will keep this section of the database current whilst also receiving result sets from the global research community. As described in this report, to enable these data sets to be effectively integrated and optimally used, HGVbaseG2P provides extensive web-based tools for result set browsing, visualization and mining.
DATABASE CONTENT AND DATA ORGANIZATION
HGVbaseG2P has evolved out of the polymorphism database HGVbase (6). In addition to extending its predecessor's scope to include phenotypes (in patient and control groups) and genotype–phenotype relationships (in the form of association study findings), HGVbaseG2P combines the best features of a database and a scientific journal, i.e. free access to structured and comprehensive result sets, along with summary-level presentation of high-quality information with full author accreditation. At the very least, the project aims to reduce hurdles to data publication and so minimize the problems of publication bias (7), and it is designed to bring together many different data sets for joint analysis thereby helping researchers identify genotyping artefacts and elucidate association signals that are population-specific or shared across related traits.
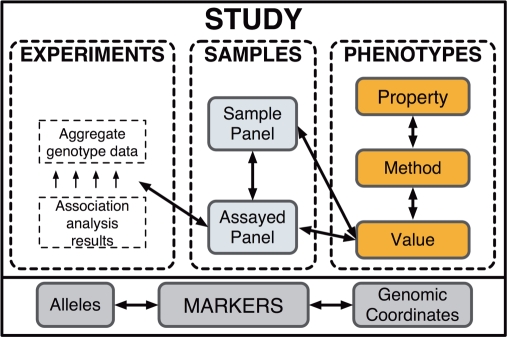
The data model underpinning HGVbaseG2P closely follows that of the new PaGE object model standard (http://www.pageom.org/). A detailed representation of the HGVbaseG2P model is available at the database website, and the principal elements of this are illustrated in Figure 1. As is shown, summary-level association data are layered on top of a foundational list of DNA variation markers. This ‘Marker’ layer currently comprises core information on all the markers presently in NCBI's dbSNP database (8) along with a direct link back to the source for each marker. Additional marker lists will be incorporated from other public databases, in particular copy-number variants which are becoming widely used in disease studies (9). Markers retain their source database identifiers and are also assigned stable HGVbaseG2P IDs. Changes to marker information in the source database are tracked and updated in HGVbaseG2P via purpose built software that appropriately handles marker and allele content alterations, mergers and deletions in a way that ensures the integrity of any existing connections to frequency or association record items.
Figure 1.
Overview data model of HGVbaseG2P. The main subdivisions of data that make up an association report are shown, with further explanation of each component provided in the body text. These components are illustrated sitting on top of a comprehensive ‘Marker’ layer which is pre-built and provides a common foundation that all data submissions are directly connected into.
The main association data layer of HGVbaseG2P comprises four principal components that emulate the main concepts used in standard literature reporting of genetic association studies, namely ‘Study’, ‘Sample’, ‘Phenotype’ and ‘Experiment’ entities.
Studies
A ‘Study’ entity sits atop, and thereby integrates, the three other main data entities that make up a single submission (‘Sample’, ‘Phenotype’ and ‘Experiment’). Each ‘Study’ entry could potentially include information items such as ‘Authors’, ‘Title’, ‘Abstract’, ‘Objectives’ and ‘Conclusions’, plus various details relating to the study design, and each will provide links to its original data source so that further information and individual level data can be requested.
Samples
HGVbaseG2P uses the ‘Sample Panel’ concept to represent a named collection of individuals that are employed in a ‘Study’—such as disease cases, or matched controls. Typically, individuals in a ‘Sample Panel’ are affected by one or more similar disease phenotypes, or have some other key metric in common (e.g. age, gender, ethnicity). Data generated by performing genotyping experiments are reported in terms of an ‘Assayed Panel’. This is a group of test subjects derived by splitting and/or merging one or more sample panels to create new collections, on the basis of some explicit criteria such as severity or subclass of disease or some environmental criteria.
Phenotypes
Phenotypes are stored in a very flexible but straightforward data structure. Whereas other databases typically use unstructured free-text descriptions to hold phenotype information, HGVbaseG2P splits phenotype information into three sub-components: (i) the ‘Phenotype Property’ which represents the character or trait investigated (e.g. nose size); (ii) the ‘Phenotype Method’ which describes how the trait was measured (e.g. nasal septum measured in centimetre to first decimal place with a ruler); and (iii) the ‘Phenotype Value’, which is a particular result produced by measuring the trait using the described method (e.g. size of nose = 1.7 cm). Identical ordinal or nominal values in groups of individuals are thereby easily represented, as are categories of disease affection status. For quantitative traits in patient groups, statistical values that describe a distribution (e.g. median, standard deviation, maximum, minimum) are stored as a series of ‘Phenotype Values’. The same data model allows phenotype thresholds to be specified and used as criteria for ‘Assayed Panel’ selection (e.g. weight greater than ‘Phenotype Value = 120 kg’).
Experiments
‘Experiments’ are individual sections of a study submission, and each is constrained to a consideration of at most one phenotype examined in a specific set of ‘Assayed Panels’ (e.g. one case and one control ‘Assayed Panel’ used to explore the role of a gene or a region or a haplotype block in causing one phenotype). Subtypes include ‘Genotyping Experiments’ (holding marker allele and genotype frequency data) and ‘Analysis Experiments’ (holding marker to trait association P-value data). The ‘Genotyping Experiments’ will become increasingly useful as a reference of marker frequencies in a range of populations as this data accumulate. Presently, however, we only permit access to aggregate allele or genotype frequency data one marker at a time, to avoid any risk of individual identification (10). The ‘Analysis Experiments’ are most central to the purpose of HGVbaseG2P, and the data for each distinct experiment may include more than one package of information based upon different statistical tests, such as an allelic trend test, a genotypic test, etc. This information will usually be initially generated as an output file from software such as PLINK (11).
EXPLORING HGVBASEG2P CONTENT
In addition to establishing a large depository of summary-level association data, an emphatic goal of HGVbaseG2P is that of providing a powerful toolkit via which the extensive database content can be explored so that new knowledge may be created.
Graphical display of association study data
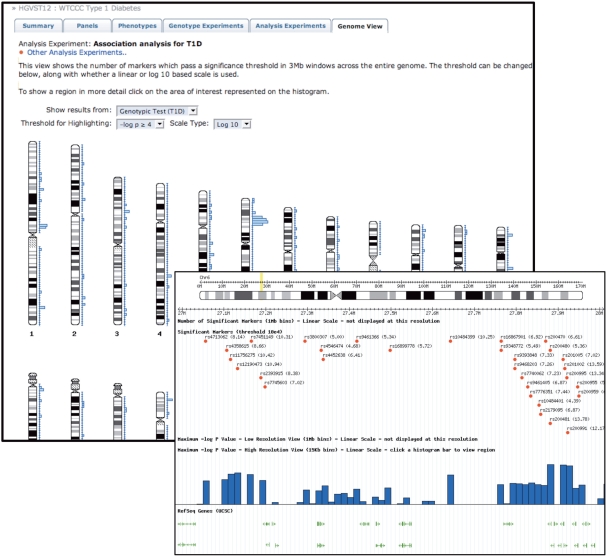
To offer an advanced visual means for accessing HGVbaseG2P content, we have utilized open-source software components from the Generic Model Organism Database project (GMOD; http://gmod.org) and configured/extended these to create novel genome-wide displays of association study findings. From these views users can then navigate onwards to far higher-resolution views provided via a region-level genome browser. This functionality is illustrated in Figure 2. These graphical displays can be reached by clicking on links provided within each ‘Analysis Experiment’ description, or by going straight to the main Genome View tab shown above every page view of a ‘Study’.
Figure 2.
HGVbaseG2P graphical display capabilities. Screenshots are provided that show data from a Type 1 diabetes disease association data set produced by the WTCCC. The histogram alongside each chromosome indicates the number (linear or logarithmic scaling options) of markers per 3 Mbp bin having a P-value that passes a tunable significance threshold. A bin with 20 significant markers is apparent at 9p21. Clicking on this region zooms in to a dynamic image (i.e. scale and position may be tailored to one's preferences) presented in a region-level browser via which the user can access optional tracks for individual marker associations, count of significant markers and P-value of most significant marker within various resolution bins, known genes details and other common annotations.
The genome browser capabilities of HGVbaseG2P have been made even more powerful still by using DAS technology—a lightweight network protocol for exchanging genome annotations (12). This enables external browsers that support DAS, such as Ensembl (13) (http://www.ensembl.org), to retrieve HGVbaseG2P data and display them as a third-party annotation track. Direct links are available from region-level genome browser views that open an Ensembl browser interface with a matching HGVbaseG2P data track already loaded.
Text-based browsing and searching
To assist users in locating particular studies of interest, we offer several alternatives. Most simply, users can enter boolean keyword search terms in search boxes provided on the home page, which trigger broad searches of the ‘Study’, ‘Phenotype’ or ‘Marker’ sections of the database. These searches return a list of all the ‘Study’ entries that contain the search term(s), with each ‘Study’ ID representing a link that will take the user to further textual and graphical information about that ‘Study’. Alternatively, one can search from the second-highest level pages in the database, which focus on ‘Study’, ‘Phenotype’ and ‘Marker’ information. These pages employ a user interface paradigm known as ‘faceted browsing’ (14). The relevant data items for these pages are listed on the right-hand side in a table or a series of panels, while the left-hand side of the page provides filtering options by which users can narrow down the set of items shown. Again, the objective is to identify one or more ‘Study’ entries of interest, to then follow their ID links to see further details on each.
Data mining
The browsing and visualization tools described above enable users to locate and then explore single ‘Study’ details. Complex data mining challenges, however, may need to query across almost all the database content and deliver complex result sets. This capability is enabled via the HGVmart tool, which is based upon another GMOD component—the BioMart system (15) (http://www.biomart.org). HGVmart functions are provided via a high-level page in the database website (derived from the standard MartView interface), as well as by means of a web service which enables programmatic query and retrieval, plus access from analysis workflow tools such as myGrid's Taverna (16) and Galaxy (17).
Gathering association data for HGVbaseG2P
Genetic association datasets are incorporated into HGVbaseG2P without any subjective evaluation of ‘importance’ of the study findings. This is central to our mission of providing a comprehensive and unbiased summary of the relationship between DNA variation and human phenotypic traits. At the time of writing (September 2008), the database is capable of holding summary-level case-control data sets for dichotomous or quantitative traits of any size or complexity. The current catalogue comprises 14 GWAS data sets spanning 12 diseases, with this initial content sourced with permission from the National Cancer Institute CGEMS project (http://cgems.cancer.gov), dbGaP, the Wellcome Trust Case-Control Consortium(WTCCC) (18) and the Broad Institute's Diabetes Genetics initiative (http://www.broad.mit.edu/diabetes/). This collection will be continually expanded by HGVbaseG2P curators who have identified several hundred additional data sets (mostly GWAS) that are now, or soon will be, publicly available. Before incorporation into HGVbaseG2P, the gathered data are checked to ensure that the utilized marker IDs are valid, that the reported alleles match known alleles for the marker in terms of sequence and DNA strand, that male and female genotype data are properly represented for X chromosome markers, and that the total association data set is complete and internally consistent.
HGVbaseG2P welcomes and encourages the direct submission of data sets from the general community, including the results of large-scale projects such as GWAS, and smaller efforts such as those designed to test individual genes, fine-map regions or replicate previously reported signals. To help with the practicality of this, we are building submission software that will guide users through the process of specifying and packaging their data, thereafter providing a means for direct submission to our curation pipeline and into the database. These developments will be supported by approaching funding agencies and scientific journals to request their help in encouraging data submissions to projects like HGVbaseG2P.
FUTURE DEVELOPMENTS
There are many ways in which HGVbase can be further developed. The underlying data model itself is quite ‘future-proof’, in that it can handle multi-marker associations, haplotype data and complex genotype classes—such as non-diploid genotypes and allele signals that may be quantitative or expressed as ratios, as needed for copy number variants. We will therefore be able to quickly incorporate these kinds of data as they become available. Ongoing work to extend the model will provide support for odds ratio data and alternative experimental designs (e.g. family-based association testing), and in conjunction with this the visualization capabilities will be suitably extended. Additional work on the genome browser is aimed at enabling researchers to view multiple sets of results simultaneously, thereby enhancing the potential for cross-study comparison and integration. Furthermore, the ability to export association data features to the Ensembl browser will be extended to integrate with the UCSC browser (19).
In all probability, an increasing number of databases like HGVbaseG2P will start to appear across the Internet in the near future (Thorisson, Muilu and Brookes, submitted). These may, like HGVbaseG2P, be designed to act as a central warehouse, or they might serve to disseminate data sets of interest to specific laboratories, institutes, consortia, societies, companies, journals or countries. It will then be highly desirable to seamlessly network these depositories into a global federated system, enabling them all to be searched from many different interfaces. This will be relatively easy to achieve if the anticipated databases all utilize common data models, such as PaGE-OM or that of HGVbaseG2P. Indeed, this concept lies at the heart of the GEN2PHEN project (http://www.gen2phen.org), of which HGVbaseG2P is a part, and to promote this our complete database implementation is freely available as open-source software. In time, this federated system can be expected to make use of increasingly powerful Grid technologies and the semantic web. HGVbaseG2P will be adapted accordingly, and our first steps in this direction are reflected in our use of the BioMart system.
SYSTEM DESIGN AND IMPLEMENTATION
Database and middleware
HGVbaseG2P is implemented as a traditional relational database using the open-source MySQL platform (http://www.mysql.com). Data import/export tools and database middleware components are all implemented in the Perl programming language, leveraging a number of open-source software packages from the Comprehensive Perl Archive Network (CPAN, http://www.cpan.org).
Website
The HGVbaseG2P web application is built using the Perl-based Catalyst MVC-framework (http://www.catalystframework.org). Genome viewer tools are based primarily on two GMOD components, the Generic Genome Browser (20) (http://gmod.org/wiki/GBrowse) and GBrowse karyotype (http://gmod.org/wiki/GBrowse_karyotype), with the Bio::DB::SeqFeature::Store feature database from the Bioperl toolkit (21) (http://www.bioperl.org) forming the back end. Web pages with faceted browsing use client-side Javascript code from the SIMILE Exhibit project (22) (http://simile.mit.edu/exhibit/). Lastly, the full-text search engine on the website was built with the open-source Xapian toolkit (http://www.xapian.org).
FUNDING
The University of Leicester; GlaxoSmithKline; and the European Community's Seventh Framework Programme (FP7/2007-2013) (grant number 200754) (the GEN2PHEN project). Funding for open access charges: European Community's Seventh Framework Programme (FP7/2007-2013).
Conflict of interest statement. None declared.
REFERENCES
- 1.Shields PG. Publication bias is a scientific problem with adverse ethical outcomes: the case for a section for null results. Cancer Epidemiol. Biomarkers Prev. 2000;9:771–772. [PubMed] [Google Scholar]
- 2.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 4.Allen NC, Bagade S, McQueen MB, Ioannidis JPA, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 5.Hulbert E, Smink L, Adlem E, Allen J, Burdick D, Burren O, Cavnor C, Dolman G, Flamez D, Friery K, et al. T1DBase: integration and presentation of complex data for type 1 diabetes research. Nucleic Acids Res. 2007;35:D742–D746. doi: 10.1093/nar/gkl933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fredman D, Munns G, Rios D, Sjoholm F, Siegfried M, Lenhard B, Lehvaslaiho H, Brookes AJ. HGVbase: a curated resource describing human DNA variation and phenotype relationships. Nucleic Acids Res. 2004;32:D516–D519. doi: 10.1093/nar/gkh111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomqvist M, Reynolds C, Katzov H, Feuk L, Andreasen N, Bogdanovic N, Blennow K, Brookes A, Prince J. Towards compendia of negative genetic association studies: an example for Alzheimer disease. Hum. Genet. 2006;119:29–37. doi: 10.1007/s00439-005-0078-9. [DOI] [PubMed] [Google Scholar]
- 8.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 10.Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J, Pearson J, Stephan D, Nelson S, Craig D. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Bakker PIWD, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yee K, Swearingen K, Li K, Hearst M. Faceted metadata for image search and browsing. CHI ′03: Proceedings of the SIGCHI conference on Human factors in computing systems. 2003;5:401–408. [Google Scholar]
- 15.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: A generic system for fast and flexible access to biological data access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Wellcome Trust Care Control Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The Generic Genome Browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, et al. The Bioperl Toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynh D, Karger D, Miller R. Exhibit: lightweight structured data publishing. WWW ′07: Proceedings of the 16th International Conference on World Wide Web. 2007:737–746. [Google Scholar]