Abstract
FLYSNPdb provides high-resolution single nucleotide polymorphism (SNP) data of Drosophila melanogaster. The database currently contains 27 367 polymorphisms, including >3700 indels (insertions/deletions), covering all major chromsomes. These SNPs are clustered into 2238 markers, which are evenly distributed with an average density of one marker every 50.3 kb or 6.6 genes. SNPs were identified automatically, filtered for high quality and partly manually curated. The database provides detailed information on the SNP data including molecular and cytological locations (genome Releases 3–5), alleles of up to five commonly used laboratory stocks, flanking sequences, SNP marker amplification primers, quality scores and genotyping assays. Data specific for a certain region, particular stocks or a certain genome assembly version are easily retrievable through the interface of a publicly accessible website (http://flysnp.imp.ac.at/flysnpdb.php).
INTRODUCTION
Drosophila melanogaster is one of the most well-studied model organisms due to its short generation time and ease of genetic manipulation. Hence, it is continuously providing major insights into biological processes which are conserved in multicellular organisms. Single nucleotide polymorphisms (SNPs) are widely used as genetic markers in mapping experiments, quantitative trait loci (QTL) analyses, population genetic or evolutionary studies, since they are frequent, mostly phenotypically neutral and molecularly defined. FLYSNPdb contains data of a polymorphism map with an unprecedented resolution of ∼50 kb between SNP markers, which is significantly higher than the density of previous Drosophila SNP maps (1–4). Polymorphisms >1 nt were also counted, including indels, which are particularly useful for genotyping assays based on PCR-product length polymorphisms [PLP; (2)] or denaturing high performance liquid chromatography [DHPLC; (5)] or also for evolutionary analyses (6,7). The map comprises SNPs from five different D. melanogaster stocks (Supplementary Table 1). Since polymorphisms in Drosophila are generally bi-allelic and randomly distributed among the utilized strains, we anticipate that most of our SNP markers can be used to discriminate almost any other pair of Drosophila stocks. FLYSNPdb is part of the FLYSNP website (http://flysnp.imp.ac.at/), which provides detailed information on the practical aspects of SNP mapping and genotyping in Drosophila (8) as well as a user guide for the database, a glossary and protocols. With this database, we want to provide a versatile SNP data resource, which is easy to use and has a user-friendly web interface.
DATA SOURCE
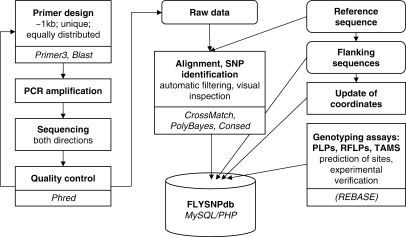
For SNP identification, we designed primer pairs to amplify fragments which are ∼1 kb long (9), equally distributed along each major chromosome arm (X, 2L, 2R, 3L and 3R), and which preferentially lie in unique, non-protein coding regions (Figure 1). Genomic DNA of up to five standard laboratory stocks per amplicon served as template: besides the wild-type stocks Canton S and Oregon R, we selected for each chromosome arm one strain that carries visible recessive markers, one stock with a Flp recombinase target (FRT) element (10) close to the centromere, and one stock with an enhancer-promoter P- (EP) element at the chromosome tip and a visible white+ marker (11) (see also Supplementary Table 1). The wild-type and FRT stocks are commonly utilized in mutagenesis screens, and the recessive marker as well as EP stocks are useful for identification of recombination events in defined chromosomal regions (2,4). PCR products were sequenced in both orientations, each using one of the amplification primers as sequencing primer. In total, >2.3 Mb (1.7%) of the 117 Mb long euchromatic region of the D. melanogaster genome were resequenced and analysed. After sequencing the PCR fragments, the Phred/Cross_match/PolyBayes software package (12–14) was used for trace quality assessment, alignment to the reference genome (strain y; cn bw sp) (15,16), and automated SNP discovery. In order to obtain high-quality data, SNPs at the first and last 75 bases of an amplicon or below Phred score 20 were omitted. In addition, ∼27% of the alignments were visually inspected [with the help of Consed 11.0 (17)], which was particularly necessary for detection of long indels (>6 bases). If multiple sequence reads from the same stock were available at one site, the allele with the highest Phred score was selected. Moreover, SNPs located at adjacent loci were considered as a single polymorphic site. Of the analysed amplicons, 86.9% contained at least one polymorphism in any of the examined stocks. The SNP positions were updated to Release 5 (FB2006_01) of the D. melanogaster genome by aligning 40 bp of the sequences (from Release 3 or 4) flanking each SNP site to the new reference sequence using Blastn (18). Prediction of restriction fragment length polymorphism (RFLP) sites was accomplished with the help of Remap [EMBOSS software suite (19,20)] and the REBASE list of commercially available restriction enzymes with cut sites ≥4 bp (21,22).
Figure 1.
Data source pipeline for SNP identification, data retrieval and curation. Software tools are displayed below each task (for references please see text); if not otherwise stated, costum-made scripts were used.
DATABASE CONTENT
The FLYSNPdb data set currently comprises >81 700 SNP alleles at 27 367 sites in 2238 amplicons of about 1 kb length (Table 1). One SNP marker contains in average 12 polymorphisms, the maximal SNP count per marker is 73. The average distance between SNP markers is 50.3 kb, a region in which one can find in average 6.6 genes [according to the FlyBase Release 5.10, FB2008_07 annotation (23)]. The biggest gap between markers is 360 kb long and lies at the tip of chromosome arm 3R, between cytological region 82A1 and 82C3. Only 169 polymorphic loci (0.6% of total SNPs) are tri-allelic, the rest is bi-allelic. A total of 13.7% (3743) of the SNPs are indels, which are up to 360 bp long, but predominantly (96.4%) <10 bp (46.6% of the indels are 1 nt long). For any given stock-pair, the average percentage of SNP markers with a sequence divergence between these two stocks is 76.6%, ranging from 35.3% to 92.0%. Furthermore, the database provides information on the molecular and cytological SNP locations for three genome assembly versions [Release 3–5; (15)], together with the 30 bp flanking sequences as additional site identification feature. For data quality assessment, PolyBayes probability scores (14), Phred trace quality scores (12), as well as the number of sequence reads per alignment are available, and manually curated SNPs are indicated. Since non-coding regions are more polymorphic, we have put our focus on non-exonic regions. If SNPs lie within an intron or exon (according to FlyBase Release 5.10), the corresponding gene name is also retrievable. In addition, information on SNP marker amplification primers is available for genotyping assays which are based on sequencing. Polymorphisms that are suitable for RFLP assays (SNPs which result in differential restriction enzyme sites) or for which verified PLP or tag-array mini-sequencing (TAMS) assays (8) are available, are also indicated, including further information like verified primers or suitable restriction enzymes.
Table 1.
Number of SNPs in FLYSNPdb, per chromosome arm and in total
| Chromosome arm | X | 2L | 2R | 3L | 3R | Total |
|---|---|---|---|---|---|---|
| SNP markers | 483 | 443 | 407 | 417 | 488 | 2238 |
| SNP sites | 4720 | 5849 | 5402 | 5993 | 5403 | 27 367 |
| Indels | 685 | 755 | 748 | 809 | 746 | 3743 |
| Alleles | 15 966 | 16 500 | 15 714 | 17 483 | 16 123 | 81 786 |
SNP sites are locations where a differential base has been identified in at least one of the stocks compared to the reference sequence or to another stock. SNP counts include number of Indels. Allele counts reflect called bases in each of the sequenced Drosophila stocks, without the alleles of the reference sequence (which are also available in FLYSNPdb).
IMPLEMENTATION, USAGE AND ACCESS
All data are organized and stored in a relational database. For increasing the speed of web queries, several summarizing tables were precomputed and put into a MySQL database which is accessed through PHP scripts.
The form on the first page asks the user to specify the chromosomal region and two stocks for which data on differential polymorphisms will be retrieved (Figure 2). The region can be indicated as molecular coordinates (position 1 – position 2 or position 1 + length) or as cytological segment (region 1 – region 2). Furthermore, it is possible to select whole chromosome arms by leaving the ‘Location’ field blank, or getting all data by selecting the ‘Browse all’ option. SNP data can be viewed as list of SNP markers (including SNP count, amplification primer sequences) or as table of SNP sites (with alleles, flanking sequences, etc.). Additional information concerning quality scores, genotyping assay suitability or coding information (genic, intronic or exonic) can be optionally selected. For users of the previous FLYSNP database version, old identifiers (ids) are retrievable and a link to this version is provided. On each query result page, sub-selections can be made by clicking on the checkboxes at the left side of each row, or by entering search parameters in the fields below each column (Figure 2). The tables are downloadable, e.g. as tab-separated text files which can be easily imported into commonly used databases or Excel spreadsheets, or as track files which can be uploaded to the FlyBase genome viewer [GBrowse; (24)]. As an additional feature, a link to FlyBase GBrowse is provided for the graphical display of the region previously specified by the user.
Figure 2.
Screenshots of FLYSNPdb input form and query result. On the first page, the user selects chromosomal region and stocks as well as different view options. On the search result page, further features such as table download or sub-queries are available.
RECENT AND FUTURE DEVELOPMENTS
The FLYSNPdb data were recently submitted to dbSNP (NCBI, Release 129; http://www.ncbi.nlm.nih.gov/projects/SNP/) so that direct linkage to the FlyBase data repository is feasible. Furthermore, sequence traces and alignments will be provided for users who would like to see the raw data for detailed quality assessment. We are open to help users with their individual needs and will implement suggestions of common use.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
European Union Fifth Framework Programme (QLRI-CT-2001-00004);Boehringer Ingelheim GmbH; Japan Society for the Promotion of Science. Funding for open access charge: IMP.
ACKNOWLEDGEMENTS
We thank the whole FLYSNP Consortium, especially Barry Dickson (IMP) for initiation and coordination of the FLYSNP project; Montserrat Aguadé and Dorcas Orengo (Univ. of Barcelona) for providing primers. We are also grateful to Cerebrum Web Consulting for initial setup of the first FLYSNP database version, Werner Kubina and Christian Brandstaetter (IMP) for IT support, Gotthold Schaffner (IMP) for sequencing and Angela Graf (IMP) for stock keeping.
REFERENCES
- 1.Teeter K, Naeemuddin M, Gasperini R, Zimmerman E, White KP, Hoskins R, Gibson G. Haplotype dimorphism in a SNP collection from Drosophila melanogaster. J. Exp. Zool. 2000;288:63–75. doi: 10.1002/(sici)1097-010x(20000415)288:1<63::aid-jez7>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 2.Berger J, Suzuki T, Senti KA, Stubbs J, Schaffner G, Dickson BJ. Genetic mapping with SNP markers in Drosophila. Nat. Genet. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- 3.Hoskins RA, Phan AC, Naeemuddin M, Mapa FA, Ruddy DA, Ryan JJ, Young LM, Wells T, Kopczynski C, Ellis MC. Single nucleotide polymorphism markers for genetic mapping in Drosophila melanogaster. Genome Res. 2001;11:1100–1113. doi: 10.1101/gr.178001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin SG, Dobi KC, St Johnston D. A rapid method to map mutations in Drosophila. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0036. RESEARCH0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nairz K, Stocker H, Schindelholz B, Hafen E. High-resolution SNP mapping by denaturing HPLC. Proc. Natl Acad. Sci. USA. 2002;99:10575–10580. doi: 10.1073/pnas.162136299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lunter G. Probabilistic whole-genome alignments reveal high indel rates in the human and mouse genomes. Bioinformatics. 2007;23:i289–i296. doi: 10.1093/bioinformatics/btm185. [DOI] [PubMed] [Google Scholar]
- 7.Lunter G, Ponting CP, Hein J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput. Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Ahlford A, Schnorrer F, Kalchhauser I, Fellner M, Viragh E, Kiss I, Syvanen AC, Dickson BJ. High-resolution, high-throughput SNP mapping in Drosophila melanogaster. Nat. Methods. 2008;5:323–329. doi: 10.1038/nmeth.1191. [DOI] [PubMed] [Google Scholar]
- 9.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 10.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 11.Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 12.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 13.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 14.Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 15.Celniker SE, Rubin GM. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 2003;4:89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- 16.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 17.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 20.Olson SA. EMBOSS opens up sequence analysis. European molecular biology open software suite. Brief Bioinform. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 21.Roberts RJ, Macelis D. REBASE–restriction enzymes and methylases. Nucleic Acids Res. 1993;21:3125–3137. doi: 10.1093/nar/21.13.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drysdale RA, Crosby MA. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]