Abstract
Over the last decade rapid progress has been made in the study of ethanol-related traits including alcohol abuse and dependence, and behavioral responses to ethanol in both humans and animal models. To collect, curate, integrate these results so as to make them easily accessible and interpretable for researchers, we developed ERGR, a comprehensive ethanol-related gene resource. We collected and curated more than 30 large-scale data sets including linkage, association and microarray gene expression from the literature and 21 mouse QTLs from public databases. At present, the ERGR deposits ethanol-related information of ∼7000 genes from five organisms: human (3311), mouse (2129), rat (679), fly (614) and worm (228). ERGR provides gene annotations and orthologs, detailed gene study information (e.g. fold changes of gene expression, P-values), and both the text and BLAST searches. Moreover, ERGR has data integration tools such as for data union and intersection, and candidate gene selection based on evidence in multiple datasets or organisms. The ERGR database is evolving with new data releases. More functions will also be added. ERGR has a user-friendly web interface with browse and search functions at multiple levels. It is freely available at http://bioinfo.vipbg.vcu.edu/ERGR/.
INTRODUCTION
Alcohol dependence, ethanol response and ethanol-related traits have been extensively studied in both humans and animal models. It is now clear that there are correlations between acute behavioral responses to ethanol and ethanol consumption or incidence of alcoholism in both animals and humans (1). In humans, alcoholism (alcohol dependence) is a common, genetically influenced complex disorder across the world. Family, twin and adoption studies demonstrated that genetic factors play a strong role in the etiology of alcoholism, accounting for 50–60% of the population variance in both men and women (2,3). Although genetic factors are important, alcoholism is a complex disease with environmental influences. Further, the architecture likely involves many genes with small effects along with environmental influences, as well as potential interactions between them. Therefore, it is a challenge to explore the molecular mechanisms underlying the genetic propensity to excessive alcohol consumption and use these for the development of new treatments for alcoholism. Many experimental strategies [linkage scan, association study, quantitative trait loci (QTLs) and microarray gene expression] have been applied in the studies of alcoholism and ethanol response in order to identify genes or chromosomal regions in both humans and model organisms (4,5).
Rapid progress in genetic studies over the past decade has identified a relatively large number of chromosomal locations or candidate genes that are linked to alcoholism, alcohol-related phenotypes and behavioral responses to ethanol (6–13). Human genetic studies have generally focused on alcohol dependence using linkage studies and association studies. The recent advances of high-throughput molecular technologies such as large-scale genotyping and DNA microarrays have greatly accelerated the generation of data used in studies searching for specific variants contributing to the genetic risk for alcoholism and ethanol response behaviors (14,15). The increasing rate of production for ethanol-related data is expected to accelerate in the near future since the cost of conducting genome-wide association studies (GWAS) is decreasing rapidly. Thus, these data provide us an unprecedented opportunity for integrating and making the wealth of results easily accessible and interpretable. So far, a few databases and computational tools, such as WebQTL (16), PhenoGen (17), WebGestalt (18) and Ontological Discovery Environment (http://ontologicaldiscovery.org/), have been developed for analyzing biological data of phenotypes and complex traits. However, the ethanol genetics research community has still lacked a comprehensive ethanol-related gene resource that presents and integrates cross-species and cross-platform data.
Here, we present such a database of ethanol-related gene resource (ERGR, http://bioinfo.vipbg.vcu.edu/ERGR/). To the best of our knowledge, it is a unique public database for ethanol-related genes. Aiming to efficiently integrate and analyze all or most of the published ethanol-related gene studies, we collected and annotated the representative large-scale ethanol-related gene datasets. These data were generated by different approaches including linkage scan, genome-wide association study, microarray expression, QTLs, retrieved from other public databases, or collected by a systematic literature search. We obtained data from the five most-studied model organisms: human, mouse, rat, fly and worm. In addition to information such as dataset description, gene annotations and gene ortholog information, the ERGR also provides tools for data integration (e.g. data union and intersection) and candidate gene selection based on multiple datasets or organisms. ERGR seeks to be a useful resource for the ethanol research community and a model database of data collection and integration for other complex diseases such as schizophrenia and Alzheimer's disease.
DATA SOURCE AND METHODS
Data collection and curation
Currently, ERGR contains ethanol-related gene data from five organisms (human, mouse, rat, fly and worm). These data were collected from different technology platforms that have been widely used in alcohol dependence or ethanol response studies: linkage scan, genome-wide association study, microarray gene expression and QTLs, or by literature search. We collected these data by the following three approaches. First, we searched publications of large-scale alcohol dependence or ethanol response studies in NCBI PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and then extracted and checked the data from these publications. Using this approach, we collected data including alcohol-related microarray gene expression studies from human or other animals, human alcohol dependence linkage studies and genome-wide association study. Specifically for the linkage data, we selected linkage regions by LOD scores and obtained the physical locations of the corresponding markers from UCSC genome browser (http://genome.ucsc.edu/) (19). Then, we retrieved the genes in the linkage regions from the Ensembl database (http://www.ensembl.org/index.html). Besides the genes in alcoholism or ethanol response studies, we also included other related genes. For example, we collected an addiction candidate gene list in which 130 genes were selected for a haplotype-based analysis of addiction (20).
Second, we extracted related data from other public databases. We obtained the mouse alcohol behavior related QTLs from the PARC (Portland Alcohol Research Center, (http://www.ohsu.edu/parc/by_phen.shtml). Only those QTLs that were marked significant and whose genomic locations could be identified were extracted. Then, we retrieved genes mapped in the QTL regions from the Mouse Genome Informatics (MGI, http://www.informatics.jax.org/) (21). Further, we searched alcohol-related genes in HuGE Navigator (http://www.hugenavigator.net/), a database of genetic associations and human genome epidemiology (22). We found nine alcohol-related phenotypes in HuGE Navigator and extracted all the genes associated with them.
Third, we performed a systematic literature search by searching the titles and abstracts of all the publications available in PubMed. We searched each protein coding gene symbol with one of the three keywords: alcohol, ethanol and alcoholism. To reduce false-positives, we manually checked those gene symbols having fewer than three letters/digits or having more than 100 hits of publications.
For each dataset, we compiled a summary description from the original data source. The summary includes the experimental method and treatment, platform, organism, tissue and phenotype, and the publication. For each gene in a dataset, more detailed information was extracted from the data source such as gene expression fold change, P-value, and tissue type.
Gene ID
We used NCBI Entrez Gene ID as the central ID for cross linking and annotation. However, specific studies used different IDs, such as gene symbol, mRNA accession number, EST accession number, clone ID, Affymetrix probe ID, Ensembl ID or UniGene ID. We applied the following three approaches to convert different IDs to gene IDs. (i) We downloaded gene2accession, gene2unigene, and gene_info files from NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/) and obtained the corresponding gene IDs for the accession numbers, UniGene IDs or gene symbols used in different studies. (ii) We used an online tool, IDConverter (http://idconverter.bioinfo.cnio.es/IDconverter.php), to convert the original IDs in the publications to gene IDs (23). (iii) When the IDs could not be converted by the above two approaches, we manually searched the NCBI databases for the gene IDs.
Gene annotation
We downloaded gene annotation files from the NCBI FTP site. Then, we extracted annotation information for the genes in our database from these files. We parsed the gene_info file to retrieve the gene information such as gene symbol, alias, full name, chromosome, genetic location and gene type. We obtained gene ontology (GO) annotations from the gene2go file downloaded from the GO website (http://www.geneontology.org/) (24). The accession numbers of the reference sequences and chromosomal location of each gene was parsed from the gene2refseq file.
Orthologs
A cross-species gene discovery and validation scheme can provide both powerful confirmation of candidate genes and mechanistic information about gene-behavior relationships. Thus, we searched orthologs of the ethanol-related genes in this database. For the nonhuman genes in our datasets, their human ortholog information was obtained and curated. We obtained human/mouse and human/rat ortholog information from MGI (ftp://ftp.informatics.jax.org/pub/reports/index.html), and human/fly and human/worm from the Inparanoid database (version 6.1, http://inparanoid.sbc.su.se/download/6.1/) (25).
DATABASE CONTENT AND ORGANIZATION
Data overview
As summarized in Table 1, we have collected and curated more than 30 genome-wide or large-scale datasets including microarray gene expression, linkage and genome-wide association studies, results of literature search and 21 mouse QTLs. At present, ERGR includes ∼7000 genes in five organisms: human (3311), mouse (2129), rat (679), fly (614) and worm (228). For rat, fly and worm, ERGR only has microarray expression data. Because of the major research interest in humans, the human data in ERGR is the most comprehensive, which includes microarray gene expression, linkage studies, genome-wide association study and literature search results.
Table 1.
Summary of the data in ERGR
| Species | Method | No. of datasets | No. of genes | Source |
|---|---|---|---|---|
| Human (Total: 3311 genes) | Microarray expression | 7 | 831 | Literature |
| Genome-wide association | 1 | 71 | Literature | |
| Linkage | 3 | 918 | Literature | |
| HuGE Navigator | 9 diseases | 203 | HuGE Navigator (http://www.hugenavigator.net/) | |
| Addiction array list | 1 | 130 | Literature | |
| Literature search | 1 | 1726 | Literature | |
| Mouse (Total: 2129 genes) | Microarray expression | 11 | 682 | Literature |
| QTL | 21 QTLs | 1568 | PARC Alcohol QTLs (http://www.ohsu.edu/parc/by_phen.shtml) | |
| Rat | Microarray expression | 6 | 679 | Literature |
| Fly | Microarray expression | 2 | 614 | Literature |
| Worm | Microarray expression | 1 | 228 | Literature |
Database organization
We used MySQL, a SQL client/sever relational open source database management system that has been commonly used in the development of biomedical databases, to store and manage the data. One table was specifically designed to store the summary descriptions of all datasets. Because the formats of the datasets generated by the different methods often varied, we managed datasets of gene expression, linkage, association or QTLs by separate tables. Annotations of gene information, gene ontology, reference sequences and ortholog information were also stored in individual tables. Dataset name, PubMed ID and Entrez gene ID are the keys to link between tables.
WEB INTERFACE
A user-friendly web interface was designed and implemented for ERGR. It is freely available at http://bioinfo.vipbg.vcu.edu/ERGR/. The user can browse and search all the data at different levels or combine the data by the integration functions.
Data browse
To help the user to browse the data easily, ERGR provides four different browsing methods: (i) by species; (ii) by method or platform such as microarray expression, linkage or association; (iii) by chromosome of a species; (iv) or through a summary page which lists all the datasets available in ERGR. A cascading style is applied for dataset browse, i.e., from dataset list to gene list, and then to gene information. By clicking the dataset name on a dataset list page it will show the dataset description and the corresponding list of genes. Selecting the gene ID will link to the gene information page, which includes gene ID, symbol and name, GO annotation, RefSeq, chromosome location, database cross links and the detailed study information (e.g. fold change of gene expression and P-value) extracted from the original ethanol studies. For example, based on the microarray dataset ‘15816859’, the fold change and P-value of ADH6 gene expression were 0.69 and 7.00 × 10−3, respectively (http://bioinfo.vipbg.vcu.edu/ERGR/geneinfo.php?id=130). Moreover, the user may find the detailed information of the studied single nucleotide polymorphisms (SNPs) via dynamic links to the NCBI dbSNP database.
Data search
ERGR provides three approaches for searching the data including text search and sequence search. First, the user may find a quick search box in the top right of the web page for searching gene ID and symbol. It supports wildcard searches such as using partial gene symbol (e.g. ADH) or using an asterisk (e.g. ADH*). Second, ERGR provides an advanced search page, on which users may combine different search terms (e.g. ID, symbol, phenotype, physical location and GO term) for a user-defined search. Third, BLAST search against the nucleotide or protein sequences of the ethanol-related genes in each organism or all the five organisms is available in ERGR.
DATA INTEGRATION
One current opportunity and challenge is the increasing large amount of public data that can be applied to the study of alcohol-related traits. Given the growth and scale of these data, efficient integration is necessary in studying a complex disorder such as alcohol dependence. Currently, ERGR provides the users data union and intersection functions for data integration; however, more functions are being developed in this ongoing project. Moreover, ERGR provides candidate gene selection results based on the evidence in multiple datasets and multiple organisms.
Data union and intersection
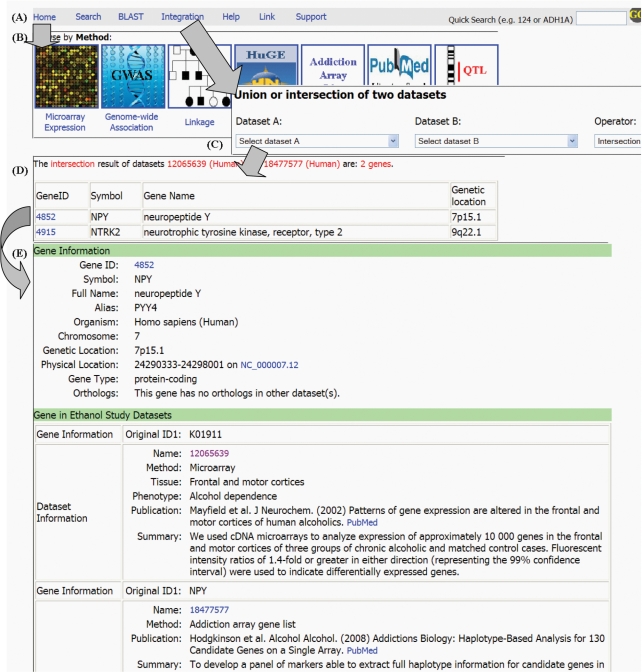
ERGR supports the union operation of any two datasets from the same organism and the intersection operation of any two datasets. There are three rules for the dataset intersection operation. First, if the two datasets to be compared are from the same organism, ERGR performs the intersection operation and outputs the results based on gene ID. Second, if the two datasets are from different organisms and one of them is from the human, ERGR uses human genes as the reference, transforms non-human genes to the human orthologs, and then performs the intersection operation based on human gene IDs. Third, if the two datasets are from two different non-human organisms, ERGR transforms genes in the both datasets to human orthologs and then performs the intersection operation based on human gene IDs. An example of data integration is shown in Figure 1.
Figure 1.
An example of data integration and gene information page. (A) Functional menu on the head of each page. (B) Data browser by method or dataset. (C) Data integration page. (D) An example of data integration. (E) Detailed information of each gene identified by the data integration (partial page).
Candidate gene selection
The candidate gene selection and prioritization function can be used to select candidate genes for follow up experimental replication or bioinformatics analysis. To make the ERGR data more useful and serve the community more effectively, ERGR provides some candidate gene selection results based on the evidence in multiple datasets either in one organism or multiple organisms. At present, ERGR contains four such candidate gene lists for ethanol response or alcoholism-related traits. The first one is a candidate gene list generated from the datasets of all organisms using human genes as reference. The non-human genes were mapped to human orthologous genes so that they could be compared systematically. There were 42 human genes or their orthologs that had evidence in more than four datasets. The other three candidate gene lists were selected from all the available datasets in one organism (human, mouse or rat) only. These candidate genes include those that have been well studied for alcohol dependence such as ADH, ALDH, GABA receptors, and NPY (4). However, the candidate gene lists also include genes with multiple lines of evidence but not so well studied such as CPE, GFAP, CRYAB, GAD1 and NTRK2, among which two [GAD1 (26) and NTRK2 (27)] had association studies reported. We found three genes (KCNJ9, GNB1 and ATP1A2) that had evidence in at least five datasets from the mouse, rat or fly but no evidence yet in any human datasets.
FUTURE PERSPECTIVES
ERGR is a unique database for ethanol-related genes and their annotations. It is freely available to public and also serves as a core data management system for the local VCU alcohol research community (VCU ARC). We will continue to collect and curate ethanol-related data, especially the genome-wide association studies. We will develop more tools that allow the users to customize their gene ranking, track their own data, and present the genes or integration results graphically.
FUNDING
The National Institute on Alcohol Abuse and Alcoholism (R21AA017437, R01AA011408, R01AA014717, U01AA016667 and U01AA016662). Funding for open access charge: R21AA017437.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We regret that numerous ethanol studies, from which the ERGR data were extracted through literature search, have not been cited in this paper because of our focus on data curation and database development. The authors would like to thank Drs. Danielle Dick, Brien Riley and Gursharan Kalsi for their valuable advice and discussions.
REFERENCES
- 1.Davies AG, Bettinger JC, Thiele TR, Judy ME, McIntire SL. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron. 2004;42:731–743. doi: 10.1016/j.neuron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br. J. Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol. Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- 4.Dick DM, Foroud T. Candidate genes for alcohol dependence: a review of genetic evidence from human studies. Alcohol. Clin. Exp. Res. 2003;27:868–879. doi: 10.1097/01.ALC.0000065436.24221.63. [DOI] [PubMed] [Google Scholar]
- 5.Schumann G, Spanagel R, Mann K. Candidate genes for alcohol dependence: animal studies. Alcohol. Clin. Exp. Res. 2003;27:880–888. doi: 10.1097/01.ALC.0000065437.18136.86. [DOI] [PubMed] [Google Scholar]
- 6.Morozova TV, Anholt RR, Mackay TF. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 2007;8:R231. doi: 10.1186/gb-2007-8-10-r231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol. Biochem. Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B, McWeeney S, Belknap J, Harrington C, Buck K, et al. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol. Clin. Exp. Res. 2004;28:1437–1448. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- 10.Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D, et al. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol. Clin. Exp. Res. 2006;30:1807–1816. doi: 10.1111/j.1530-0277.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- 11.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am. J. Med. Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- 12.Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J. Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J. Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman P, Tabakoff B. Gene expression in animals with different acute responses to ethanol. Addict. Biol. 2005;10:63–69. doi: 10.1080/13556210412331308985. [DOI] [PubMed] [Google Scholar]
- 15.Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B:844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- 17.Bhave SV, Hornbaker C, Phang TL, Saba L, Lapadat R, Kechris K, Gaydos J, McGoldrick D, Dolbey A, Leach S, et al. The PhenoGen informatics website: tools for analyses of complex traits. BMC Genet. 2007;8:59. doi: 10.1186/1471-2156-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, et al. Addictions Biology: Haplotype-Based Analysis for 130 Candidate Genes on a Single Array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 23.Alibes A, Yankilevich P, Canada A, Diaz-Uriarte R. IDconverter and IDClight: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berglund AC, Sjolund E, Ostlund G, Sonnhammer EL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh el W, Lane HY, Chen CH, Chang PS, Ku LW, Wang KH, Cheng AT. Glutamate decarboxylase genes and alcoholism in Han Taiwanese men. Alcohol. Clin. Exp. Res. 2006;30:1817–1823. doi: 10.1111/j.1530-0277.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- 27.Xu K, Anderson TR, Neyer KM, Lamparella N, Jenkins G, Zhou Z, Yuan Q, Virkkunen M, Lipsky RH. Nucleotide sequence variation within the human tyrosine kinase B neurotrophin receptor gene: association with antisocial alcohol dependence. Pharmacogenomics J. 2007;7:368–379. doi: 10.1038/sj.tpj.6500430. [DOI] [PMC free article] [PubMed] [Google Scholar]