Abstract
Plant hormones are small organic molecules that influence almost every aspect of plant growth and development. Genetic and molecular studies have revealed a large number of genes that are involved in responses to numerous plant hormones, including auxin, gibberellin, cytokinin, abscisic acid, ethylene, jasmonic acid, salicylic acid, and brassinosteroid. Here, we develop an Arabidopsis hormone database, which aims to provide a systematic and comprehensive view of genes participating in plant hormonal regulation, as well as morphological phenotypes controlled by plant hormones. Based on data from mutant studies, transgenic analysis and gene ontology (GO) annotation, we have identified a total of 1026 genes in the Arabidopsis genome that participate in plant hormone functions. Meanwhile, a phenotype ontology is developed to precisely describe myriad hormone-regulated morphological processes with standardized vocabularies. A web interface (http://ahd.cbi.pku.edu.cn) would allow users to quickly get access to information about these hormone-related genes, including sequences, functional category, mutant information, phenotypic description, microarray data and linked publications. Several applications of this database in studying plant hormonal regulation and hormone cross-talk will be presented and discussed.
INTRODUCTION
Over the past century, extensive physiological and morphological studies have demonstrated that almost every aspect of plant growth and development is regulated by a small set of organic molecules referred to as plant hormones (1,2). In the recent decades, a great effort had been made by using the model plant Arabidopsis thaliana to uncover the genetic basis of plant hormonal regulation. Normally, a relatively simple and specific morphological response to a given type of hormone would be used as a screening phenotype to isolate genetic mutations that result in reduced or enhanced sensitivity to such hormone. Dozens of mutants that show altered response to certain type of hormone have been isolated and characterized for their involvement in plant hormone actions. As a result, a large number of genes that participate in various plant hormone responses, including hormone biosynthesis, metabolism, transport, perception and signal transduction, have been identified and characterized by means of forward genetic and reverse genetic approaches (3). However, there is so far not a collective database that could provide a systematic and comprehensive description on morphological phenotypes regulated by plant hormones, as well as regulatory genes participating in numerous plant hormone responses.
It has been found that many biological processes are coordinately regulated by multiple hormones. Recent physiological and genetic studies together with the large-scale analyses of microarray data indicated that there exist a wide range of cross-talks among different classes of plant hormones (4). The combination of these signals controls plant growth, development and response to myriad biotic and abiotic stresses in a complex manner (4,5). For example, ethylene and auxin can regulate a number of common processes including primary root elongation, root hair formation, hook formation, leaf epinasty and abscission (6). However, the molecular basis of many cross-talks among different hormones and signals remain poorly understood. It is thus necessary to summarize on how many types of plant hormones and in particularly what genes are involved in each kind of hormone-regulated phenotypes. This collection would certainly be of great help to plant researchers that are interested in deciphering the molecular code underlying those cross-talks among myriad plant hormone actions.
Here, we report a database that collects 1026 Arabidopsis genes related to the actions of eight plant hormones, including auxin (IAA), gibberellin (GA), cytokinin (CK), abscisic acid (ABA), ethylene (ET), jasmonic acid (JA), salicylic acid (SA) and brassinosteroid (BR). The database offers comprehensive information about plant hormone-related genes and plant hormone-regulated morphological processes. Meanwhile, a phenotype ontology is developed to systematically and precisely describe myriad hormone-regulated morphological processes using standardized vocabularies. Users can quickly get access to information about most, if not all, hormone-related genes reported in literatures from the web interface of the Arabidopsis hormone database (AHD) (http://ahd.cbi.pku.edu.cn). This user-friendly website allows searching for hormone-related genes and mutants by names, sequences, free texts, response hormones and mutant phenotypes.
DATABASE CONTENT
Collection of plant hormone-related genes
We have identified Arabidopsis genes related to plant hormone functions (hormone-related genes) by following ways. First, by extensively reviewing literature of more than 330 papers published before August 2008, we collected 302 genes that have been shown to participate in various aspects of hormone functions by genetic evidence (mutant analysis or transgenic over-expression studies). Among them, 282 genes were supported by mutant analysis, 36 genes were derived from transgenic studies (Table 1), and 16 genes were supported by both mutant analysis and transgenic studies. We classified these genes into distinct functional categories, including hormone biosynthesis, hormone transport, hormone metabolism, hormone perception, hormone signal transduction and other aspects of hormone responses (Table 2). Second, we developed a Perl script to extract genes mapped to 72 gene ontology (GO) terms that are related to hormone actions (GO terms are listed in Table S1) from TAIR Arabidopsis gene ontology annotation. We collected totally 923 genes that are implicated in plant hormonal regulation in this manner (Table 1), of which, 199 genes were supported by genetic evidence. Combining the two sources of data together, we identified a total of 1026 Arabidopsis genes related to plant hormone actions (Table 1). This number might change after the database is constantly updated in the future.
Table 1.
Hormone-related genes in the AHD
| Hormone | No. of genes (supporting evidence) |
|||
|---|---|---|---|---|
| Genetic study |
Gene Ontology annotation | All | ||
| Mutant | Transgenic | |||
| Auxin | 59 | 2 | 323 | 335 |
| Gibberellin | 25 | 10 | 146 | 149 |
| Cytokinin | 18 | 8 | 72 | 80 |
| Abscisic acid | 78 | 1 | 252 | 289 |
| Ethylene | 31 | 5 | 156 | 166 |
| Jasmonic acid | 33 | 7 | 152 | 171 |
| Salicylic acid | 22 | 2 | 116 | 135 |
| Brassinosteroid | 35 | 2 | 43 | 58 |
| ALL hormone | 282 | 36 | 923 | 1026 |
Table 2.
Functional categories of hormone-related genes
| Hormone | Functional categories | No. of all genesa |
|---|---|---|
| Auxin | Hormone biosynthesis | 9 |
| Auxin | Hormone metabolism | 3 |
| Auxin | Hormone receptor | 4 |
| Auxin | Hormone signal transduction | 33 |
| Auxin | Hormone transport | 12 |
| Gibberellin | Hormone biosynthesis | 15 |
| Gibberellin | Hormone metabolism | 2 |
| Gibberellin | Hormone receptor | 3 |
| Gibberellin | Hormone signal transduction | 9 |
| Cytokinin | Hormone biosynthesis | 1 |
| Cytokinin | Hormone metabolism | 4 |
| Cytokinin | Hormone receptor | 4 |
| Cytokinin | Hormone signal transduction | 15 |
| Cytokinin | Hormone transport | 2 |
| Abscisic acid | Hormone biosynthesis | 9 |
| Abscisic acid | Hormone metabolism | 2 |
| Abscisic acid | Hormone receptor | 3 |
| Abscisic acid | Hormone signal transduction | 65 |
| Abscisic acid | Plant development | 2 |
| Ethylene | Hormone biosynthesis | 6 |
| Ethylene | Hormone receptor | 5 |
| Ethylene | Hormone response | 2 |
| Ethylene | Hormone signal transduction | 19 |
| Jasmonic acid | Hormone biosynthesis | 12 |
| Jasmonic acid | Hormone receptor | 1 |
| Jasmonic acid | Hormone signal transduction | 24 |
| Salicylic acid | Hormone biosynthesis | 1 |
| Salicylic acid | Hormone metabolism | 3 |
| Salicylic acid | Hormone signal transduction | 18 |
| Salicylic acid | Hormone transport | 1 |
| Brassinosteroid | Hormone biosynthesis | 12 |
| Brassinosteroid | Hormone metabolism | 2 |
| Brassinosteroid | Hormone receptor | 3 |
| Brassinosteroid | Hormone signal transduction | 18 |
aGenes supported by genetic evidence.
Description of plant hormone-regulated phenotypes
For 302 plant hormone-related genes that have mutant or transgenic plant information, we describe phenotypes of these mutants and transgenic plants by comprehensive literature review.
Integration of expression data of hormone-regulated genes
High-throughput gene expression profiling allows a global analysis of transcriptional regulation in hormone responses. Gene expression arrays have been used to examine the RNA levels of Arabidopsis genes in response to exogenous treatments of a given hormone, or in various hormone-response mutants in Arabidopsis. A large collection of hormone-regulated genes have been identified in a series of published studies and are available to public domain (all references listed in the website). In the AHD, we integrated the expression data of the hormone-regulated genes derived from the supplementary data of numerous published papers. All expression levels of hormone-regulated genes are converted to and presented in log2 ratio to facilitate the comparison. The description of Materials and Methods for the experiments, the calculation of ratio, type of microarray (most of them are AFFYMETRIX chips), URLs of the original data and the links to the corresponding papers are enclosed in the general information for each microarray experiment.
WEB INTERFACE
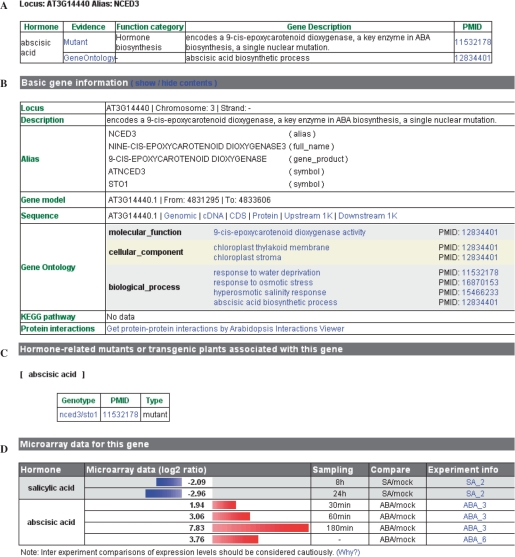
The AHD provides user-friendly interface to display information. The information of hormone-related genes contains four sections (Figure 1). The first section indicates the type of hormone(s) related to the gene. Additional information in this section includes supporting evidence, functional categories, gene description and PubMed ID of the associated references (Figure 1A). The second section describes the basic gene information retrieved from other databases, including locus name, gene description, gene alias, gene model, gene sequences, gene ontology annotation from TAIR version 7 database (7), metabolic pathway annotation from KEGG database (8) and protein–protein interaction (PPI) data (Figure 1B). The third section lists the reported mutants (including transgenic plants) associated with the given gene (Figure 1C). Click on the links to the listed mutants will allow access to their respective phenotypes. The detailed information about the linked mutants is described below. The fourth section displays a graphic view of gene expression data if this gene is differentially regulated by one or multiple types of hormones based on the previously published microarray studies (Figure 1D).
Figure 1.
Screenshot of gene information. The information of a hormone-related gene contains four sections, including (A) response hormone(s), (B) basic gene information retrieved from TAIR and KEGG, (C) associated mutants and (D) expression data from microarray experiments.
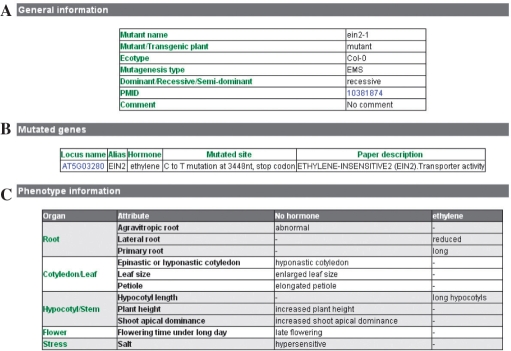
As to the mutant information, each mutant contains three sections: general information, gene(s) associated with the mutant and description of mutant phenotypes (Figure 2). General information includes the mutant's ecotype, mutagenesis type and genetic feature (Figure 2A). The genotype information lists the mutated genes and mutated site for the given mutant (Figure 2B). Click on the links to the genes will allow access to detailed gene information (Figure 1). We have developed a standardized classification system to describe phenotypic traits of mutants identified in plant hormone research (Figure 2C). Phenotypic traits are classified into seven classes: root, cotyledon/leaf, hypocotyl/stem, flower, silique/seed, embryo and stress. These seven classes are further categorized into 63 subclasses (Figure 3). For instance, class ‘root’ can be further divided into six subclasses: primary root, lateral root, root hairs, agravitropic root, swollen primary roots or lateral roots and other. Most root-related phenotypic traits reported in literatures can be included into the first five subclasses. Occasionally, if the root phenotypic traits are difficult to be classified into any of the above five subclasses, we categorize them into a subclasses designated as ‘other’ (Figure 3). All phenotypic traits of mutants are described by this standardized classification system.
Figure 2.
Screenshot of mutant information. The information of a mutant consists of three sections, including (A) general information, (B) genes corresponding to the mutant and (C) mutant phenotype information.
Figure 3.
Screenshot of phenotype ontology. The phenotype ontology consists of 7 major classes and 63 subclasses. Each phenotype can be classified into a provided subclass. Mutants with a given phenotype and genes associated with those mutants can be accessed through the hyperlinks in phenotype ontology.
We have developed powerful search system to help users retrieve information. For gene information, users can search genes by gene name, gene description or responding hormones. We also provide NCBI BLAST to help users to search genes by sequences. For mutant information, users can search mutants by mutant name and hormones. Phenotype search allow users to acquire all mutants that have selected phenotypic traits, as well as genes associated with these mutants. For microarray data, users can search microarray experiments and retrieve expression data by type of hormone treatments and plant organ types. A detailed tutor on how to use the search system is provided on the website (http://ahd.cbi.pku.edu.cn/help).
THE APPLICATION OF AHD
Application of phenotype ontology
Hormone-regulated phenotypes and their associated genes
Although different types of hormones might regulate distinctive aspects of plant growth and developmental processes, a common phenotypic trait could be controlled by multiple hormones (4). To systematically examine the effect of different hormones on any given phenotypic traits, we determine the number of genes and the types of hormones that regulate each subclass of phenotype. Among 60 phenotype subclasses we examined, 51 are regulated by multiple hormones (Table 3). Phenotypic traits that are related to trichome branching, stomata density, nonphototropic hypocotyls, gynoecium protrusion, no embryonic root, embryogenesis stage, cold, drought and osmosis are regulated by a single type of hormone according to literatures (Table 3). Therefore, our analysis support the notion that, although generally considered to play distinctive roles in regulating various aspects of plant life cycle, different types of hormones also act together to modulate the same biological processes in a complex manner.
Table 3.
The summary of hormone-regulated phenotypes
| Phenotypic traits | No. of gene | No. of hormone | List of hormone |
|---|---|---|---|
| Root | 148 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Primary root | 122 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Lateral root | 67 | 6 | IAA, CK, ABA, ET, JA, BR |
| Root hairs | 36 | 6 | IAA, CK, ABA, ET, JA, BR |
| Agravitropic root | 27 | 5 | IAA, ABA, ET, JA, BR |
| Swollen primary roots or lateral roots | 13 | 4 | IAA, ET, JA, BR |
| Cotyledon/Leaf | 162 | 8 | IAA, GA, CK, ABA, ET, JA, SA, BR |
| Cotyledon number | 11 | 5 | IAA, GA, ABA, ET, JA |
| Epinastic or hyponastic cotyledon | 28 | 5 | IAA, CK, ABA, ET, BR |
| Vasculature pattern | 26 | 5 | IAA, ABA, ET, JA, BR |
| Leaf size | 83 | 8 | IAA, GA, CK, ABA, ET, JA, SA, BR |
| Rumpled or serrated leaves | 29 | 5 | IAA, ABA, ET, JA, BR |
| Hyponastic or epinastic leaves | 41 | 6 | IAA, GA, ABA, ET, JA, BR |
| Rounded or narrow leaves | 46 | 5 | IAA, ABA, ET, JA, BR |
| Trichome branching | 1 | 1 | GA |
| Stomata aperture | 24 | 3 | ABA, ET, BR |
| Stomata density | 1 | 1 | ABA |
| Leaf senescence | 13 | 4 | CK, ABA, ET, BR |
| Leaf color | 63 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Petiole | 52 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Leaf growth in dark | 7 | 4 | IAA, ABA, ET, BR |
| Hypocotyl/stem | 145 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Hypocotyl length | 103 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Shoot apical hook | 38 | 5 | IAA, GA, ABA, ET, BR |
| Agravitropic hypocotyls | 11 | 2 | IAA, ABA |
| Nonphototropic hypocotyls | 6 | 1 | IAA |
| Plant height | 95 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Shoot apical dominance | 50 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Internode length | 35 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Flower | 97 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Flowering time under long day | 30 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Flowering time under short day | 13 | 2 | GA, ABA |
| Flower size | 16 | 5 | IAA, ABA, ET, JA, BR |
| Sepal | 15 | 4 | IAA, ET, JA, BR |
| Petal | 16 | 4 | IAA, ET, JA, BR |
| Stamen | 26 | 6 | IAA, GA, ABA, ET, JA, BR |
| Carpel | 13 | 5 | IAA, GA, ET, JA, BR |
| Gynoecium protrusion | 2 | 1 | IAA |
| Anther development | 20 | 5 | IAA, ABA, ET, JA, BR |
| Pollen development | 24 | 6 | IAA, GA, ABA, ET, JA, BR |
| Fertility | 71 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Silique/Seed | 128 | 7 | IAA, GA, CK, ABA, ET, JA, BR |
| Silique length | 44 | 6 | IAA, GA, ABA, ET, JA,BR |
| Silique shape | 10 | 4 | IAA, ABA, ET, BR |
| Germination rate | 67 | 5 | IAA, GA, ABA, ET, JA |
| Loss of seed dormancy | 21 | 4 | GA, CK, ABA, BR |
| Seed yield | 45 | 6 | IAA, GA, ABA, ET, JA, BR |
| Seed size | 5 | 4 | IAA, CK, JA, BR |
| Seed shape | 3 | 2 | ABA, JA |
| Embryo | 21 | 6 | IAA, GA, ABA, ET, JA, BR |
| Defective embryo development | 15 | 4 | IAA, ABA, JA, BR |
| No embryonic root | 9 | 1 | IAA |
| Embryo lethality | 8 | 2 | IAA, ABA |
| Embryogenesis stage | 2 | 1 | IAA |
| Stress response | 88 | 6 | IAA, GA, ABA, ET, JA |
| Salt | 23 | 3 | GA, ABA, ET |
| Cold | 3 | 1 | ABA |
| Drought | 12 | 1 | ABA |
| Pathogen | 38 | 3 | ET, JA |
| Osmosis | 13 | 1 | ABA |
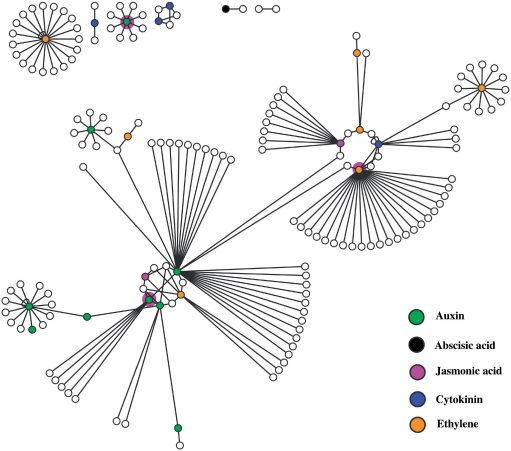
In order to determine how different hormones coordinate the regulation of a common phenotypic trait, we further analyzed a phenotypic trait that was reported to be regulated by multiple hormones: primary root elongation. Based on our database search, we have identified 120 mutants that show a longer primary root length than wild type in a normal or hormone-treated condition. These mutants correspond to a total of 63 genes that regulate this phenotype. Among them, 12 genes are related to CK responses, 24 are ET-related genes, 2 are ABA-related, 18 are auxin-related, 11 are JA-related, and 1 is for GA and 1 for BR. To investigate the molecular mechanism of how these genes function coordinately to regulate primary root elongation, we tested whether their encoding products show PPI. Of 63 proteins, 23 can be mapped into protein-protein interaction networks via the web server of the Arabidopsis interactions viewer (http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi). And 17 out of 23 hormone-related proteins are interconnected by direct interactions or through intermediate protein(s) in a single network (Figure 4, Table S2). Interestingly, only three genes are involved in response to multiple hormones, whereas 14 others are related to a single type of hormone. This result implies that different hormones regulate a common process likely by sharing an interconnected regulatory network, but not by the same genes.
Figure 4.
PPI network of genes involved in regulating primary root length. The circles represent proteins and the lines indicate PPIs. The colored circles represent hormone-related proteins when mutated would cause a longer primary root phenotype. Different colors indicate different hormone classes. The uncolored circles represent proteins that show direct interaction with hormone-related proteins.
Co-regulation of different phenotypes
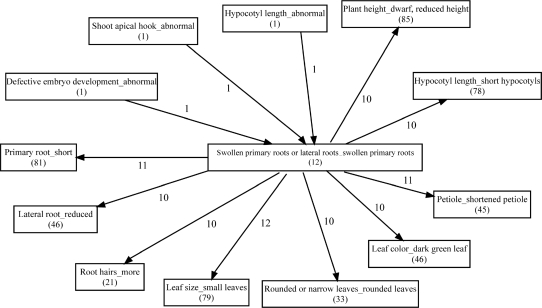
Based on our database analysis, we found that the majority of phenotypic traits are regulated by a group of genes involved in multiple hormone responses (Table 3). We then surveyed whether different phenotypes could be regulated by the same group of genes. We have identified 406 pairs of phenotypic traits, in which over 80% of associated genes in one trait are shared by the other trait (Table S3). For example, 12 genes have been shown to regulate swollen primary roots. Of the 12 genes, we found that all 12 genes regulate small leaves, 11 genes regulate primary root length and shortened petiole, 10 genes regulate either reduced plant height, short hypocotyls, dark green leaf, rounded leaves, more root hairs or reduced lateral root, respectively (Figure 5). This result demonstrates that some phenotypic traits are highly related, suggesting that these correlated phenotypes might be physiologically or ecologically interconnected, or share the same origin in the evolution course.
Figure 5.
Overlap of regulatory genes among different phenotypes. Each rectangle represents a phenotype. The number of genes regulating that phenotype is shown in parenthesis. The number on the arrow, which arrowhead points to the large geneset and arrowend points to the small geneset, represents the overlap of genes regulating two different phenotypes.
Hormone-related genes involved in multiple hormone functions
One mechanism of hormone cross-talks is to share key regulatory components in multiple hormone response pathways. We found that almost any two types of hormones share more or less regulatory genes, except for BR and JA, BR and SA, and CK and JA (Table S4). We then explored how many hormone-related genes are involved in multiple hormone responses. Of the total 1026 hormone-related genes supported by either genetic evidence or GO annotation evidence, we found that ∼17% of them participate in the function of more than one type of hormones (Figure S1, Table S5). At the extreme situation, we found 21 genes that can respond to six hormones. Interestingly, all 21 genes encode transcription factors, 10 for MYB proteins and 11 for MYB-related proteins, suggesting that transcriptional regulation might be the key integration points in plant hormonal interactions.
DISCUSSION
The AHD integrates a large volume of literature data based on mutant studies, transgenic analysis, gene ontology annotation and microarray studies. A pronounced characteristic of this database is that we have developed a new set of standardized vocabularies to describe the phenotypic traits of a comprehensive collection of mutants identified in plant hormone research. It is easy to search for mutants and genes related to any given phenotype from this phenotype ontology. To date, several databases for Arabidopsis mutants are available (9–12), among which three databases have no mutant phenotype description (9,11,12) and one developed its own phenotypic classification system (10). In comparison with previously developed databases, the AHD phenotype ontology system has several distinctions and virtues: first, this system concentrates on Arabidopsis hormone-related mutants, and such a database had not been developed before. Second, this system integrates phenotypic traits derived from a large number of published studies supported by sound genetic evidence. Third, this system contains a more complete list of phenotype subclasses, and describes phenotypic traits in more details for hormone-related mutants compared with other databases (10).
To our knowledge, this database represents the first effort to systematically collect a wide range of published results, and contains the most comprehensive information on both hormone-related phenotypes and Arabidopsis genes regulating these phenotypes. Users can quickly get access to information about most, if not all, genes reported in literatures that regulate a given hormone function, including gene sequences, functional category, mutant information, phenotypic description, microarray data and linked publications. The phenotype ontology database provides a novel route to study plant hormone actions and interactions. It has been shown that many biological processes are regulated by multiple hormones that interplay in different ways, although the molecular basis of such interactions is not known. By use of this AHD database, we have conducted analyses on the interaction among multiple hormone pathways in regulating primary root elongation. Our analysis suggests that different phytohormones control this common trait probably by sharing a physically interconnected regulatory network, but not by sharing the same genes. We have discovered that certain groups of phenotypes are highly related due to the large overlap of their regulatory genes. We have also identified a class of multiple-hormone responsive MYB and MYB-like transcription factors that might mediate responses to as many as six hormones. These analyses demonstrate that our database is valuable in unraveling molecular mechanisms of plant hormone cross-talks in regulating various biological processes.
Our database aims at providing a comprehensive and standardized resource for Arabidopsis hormone research. Each type of hormone has one or two experts to curate the database in a user-compatible manner, and to update the information in a timely manner. It is our belief that the database will benefit plant hormone researchers all over the world. Users can also help to improve the database via our interactive platform (http://ahd.cbi.pku.edu.cn/help.php). All useful information and comments will be properly considered and quickly integrated into the database by corresponding curators for each hormone.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation, China (NSFC30625003 and ED20060047 to H.G.). Funding for open access charge: 111 project.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Qiyao Li for web decoration, Fengying An and Qiong Zhao for helpful discussion.
REFERENCES
- 1.Davies PJ. The Netherlands: Kluwer Academic Publishers, Dordrecht; 1995. The Plant Hormones: Their Nature, Occurrence and Functions. [Google Scholar]
- 2.Bishopp A, Mahonen AP, Helariutta Y. Signs of change: hormone receptors that regulate plant development. Development. 2006;133:1857–1869. doi: 10.1242/dev.02359. [DOI] [PubMed] [Google Scholar]
- 3.Alonso JM, Ecker JR. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat. Rev. Genet. 2006;7:524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- 4.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 5.Seki M, Satou M, Sakurai T, Akiyama K, Iida K, Ishida J, Nakajima M, Enju A, Narusaka M, Fujita M, et al. RIKEN Arabidopsis full-length (RAFL) cDNA and its applications for expression profiling under abiotic stress conditions. J. Exp. Bot. 2004;55:213–223. doi: 10.1093/jxb/erh007. [DOI] [PubMed] [Google Scholar]
- 6.Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Rosso MG, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 2007;35:D874–D878. doi: 10.1093/nar/gkl753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, Takabe H, Sakurai T, Akiyama K, Hirayama T, et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- 11.Pan X, Liu H, Clarke J, Jones J, Bevan M, Stein L. ATIDB: Arabidopsis thaliana insertion database. Nucleic Acids Res. 2003;31:1245–1251. doi: 10.1093/nar/gkg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya A, Mizukado S, Sakurai T, Shinozaki K. A resource of 5,814 dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol. 2005;46:1149–1153. doi: 10.1093/pcp/pci112. [DOI] [PubMed] [Google Scholar]