Abstract
‘miR2Disease’, a manually curated database, aims at providing a comprehensive resource of microRNA deregulation in various human diseases. The current version of miR2Disease documents 1939 curated relationships between 299 human microRNAs and 94 human diseases by reviewing more than 600 published papers. Around one-seventh of the microRNA–disease relationships represent the pathogenic roles of deregulated microRNA in human disease. Each entry in the miR2Disease contains detailed information on a microRNA–disease relationship, including a microRNA ID, the disease name, a brief description of the microRNA–disease relationship, an expression pattern of the microRNA, the detection method for microRNA expression, experimentally verified target gene(s) of the microRNA and a literature reference. miR2Disease provides a user-friendly interface for a convenient retrieval of each entry by microRNA ID, disease name, or target gene. In addition, miR2Disease offers a submission page that allows researchers to submit established microRNA–disease relationships that are not documented. Once approved by the submission review committee, the submitted records will be included in the database. miR2Disease is freely available at http://www.miR2Disease.org.
INTRODUCTION
MicroRNAs are a class of endogenous single-stranded small noncoding RNAs that negatively regulate gene expression (1). MicroRNAs control mRNA degradation and/or translation inhibition through binding to complementary sites in the 3′-untranslated regions of target genes (2). In the past decade, hundreds of microRNAs have been identified in mammalian cells (3,4). Evidence suggests that microRNAs play critical roles in multiple biological processes, including cell cycle control, cell growth and differentiation, apoptosis, embryo development and so on (5–8).
In recent years, dozens of microRNA-related database systems have been developed. miRBase (9), miRGator (10) and miRGen (11) aim at providing complete repositories for microRNA annotation and nomenclature. TarBase (12), microRNAMap 2.0 (13) and microRNA.org (14) collect experimentally validated and/or computationally predicted microRNA–target relationships; microRNA.org also provides a collection of microRNA expression profiles in different tissues. Such database systems offer great resources in investigating the function of microRNA in gene regulation. In addition, several computational algorithms and web-based programs have been developed to computationally predict target genes/sites of microRNAs, such as TargetScan (15), PicTar (16), RNAhybrid (17) and PITA (18).
Accumulating evidence indicates that microRNAs play crucial roles in human disease development, progression, prognosis, diagnosis and evaluation of treatment response (19–24). Genome-wide association studies demonstrated that many human microRNA genes locate at genomic regions linked to cancer (25,26). Moreover, a recent study found that the absolute expression levels of many microRNAs were reduced significantly in tumors (27). The same study also revealed that using the expression levels of 217 microRNAs were more effective in cancer classification than mRNA microarrays containing more than 16 000 protein-coding genes (27). All these evidences support the strong necessities in understanding the function of microRNA in disease development.
In addition to these genome-wide studies, correlations between single microRNA deregulation and occurrences of human disease have been reported. Cimmino et al. (28) demonstrated that both miR-15a and miR-16-1 negatively regulate BCL2 at a post-transcriptional level, which induces apoptosis in a leukemic cell line model. Yang et al. (29) found that miR-1 overexpression slowed cardiac conduction and depolarized the cytoplasmic membrane by post-transcriptionally repressing KCNJ2 and GJA1, which accounted at least in part for its arrhythmogenic potential. Hitherto, over 100 review papers have been published in summarizing the relationship between microRNA deregulation and dozens of diseases (30–36). Despite these developments, detailed information on microRNA–disease relationships are scattered in literatures and there is no online repository for these microRNA–disease relationships. Therefore, we develop a manually curated database entitled ‘miR2Disease’ (http://www.miR2Disease.org), which provides a comprehensive resource of microRNA deregulation in various human diseases.
DATA COLLECTION AND DATABASE CONTENT
Initial entries describing the relationships between microRNA deregulation and occurrences of human disease are collected manually. We searched the PubMed database (37) with a list of keywords, such as ‘microRNA disease’, ‘miRNA disease’, ‘microRNA cancer’, ‘miRNA cancer’, etc. In the current release of miR2Disease, more than 600 literatures were reviewed, and 1939 curated relationships between 299 human microRNAs and 94 human diseases were documented. In the miR2Disease system, the disease terminologies were organized based upon a controlled medical vocabulary (Disease Ontology: http://diseaseontology.sourceforge.net/), which utilizes the Unified Medical Language System (UMLS) as source vocabulary (38). Such organization provides substantial advantage in terms of search and analysis. Each entry in the database contains detailed information on a microRNA–disease relationship, including a microRNA ID, the disease name, a brief description of the microRNA–disease relationship, an expression pattern of microRNA (upregulated or downregulated) in the disease state, the detection method used to derive microRNA expression pattern (microarray, northern blot, qRT-PCR, etc.), the experimentally validated target gene(s) extracted from the corresponding references and directly derived from TarBase and a literature reference. miR2Disease provides a user-friendly interface for an easy query of each entry by microRNA ID, disease name or experimentally validated target genes. In addition, convenient links are provided to other microRNA databases, including microRNA sequence and annotation in miRBase, experimentally validated microRNA target genes in TarBase and computationally predicted microRNA target genes in TargetScan, miRanda and PicTar. A hyper link to the literature reference in the NCBI PubMed database (37) together with an official PubMed ID and complete citation are also provided. In miR2Disease, around one-seventh of the microRNA–disease relationships represent the pathogenic roles of deregulated microRNA in human disease.
USER INTERFACE
Search page
miR2Disease provides a search engine to query detailed information on each microRNA–disease relationship documented in the database. Users can query the database through microRNA ID, disease name or target genes.
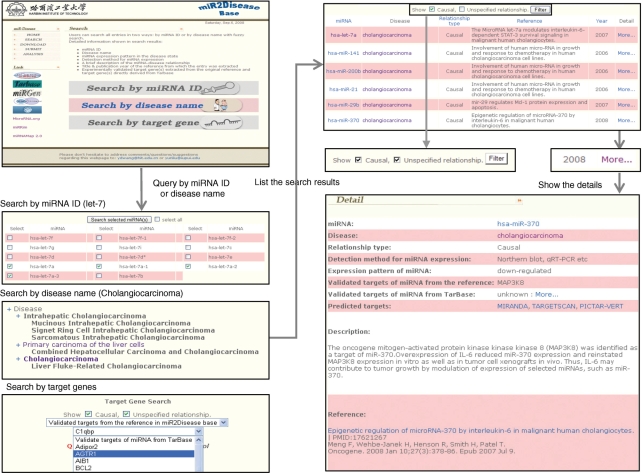
miR2Disease offers a fuzzy search function. Combining with controlled disease vocabulary (disease ontology), fuzzy search functionality allows users to retrieve meaningful microRNA–disease relationship information without knowing the exact disease name documented in the database. Once certain disease name is received as a query term, the system first searches all the disease terminologies that contain the query words. The matching disease terminology will be listed in the context of a disease tree, which contains its ancestor nodes as well as all of its subcategories. In the matching disease tree, the categories that contain query terminologies are highlighted as bold, and a hyperlink is created for every category that contains documented microRNA–disease relationships. Through these hyperlinks, users can retrieve all the microRNA entries that are related to the selected disease terminology. In the search results page, clicking the ‘more …’ link at the end of the entry will lead to detail information of this relationship (Figure 1).
Figure 1.
A schematic workflow of miR2Disease.
Similar to search by disease terminology, the nomenclatures of microRNAs could be complicated and confusing. It is not unusual that the original publication did not provide enough information on the exact microRNA in a large microRNA family. For instance, some publications simply report let-7 is related to adenoma, while others conclude that the expression level of let-7a-3 is downregulated in breast cancer. Therefore, through the fuzzy search function, miR2Diesease allows queries of microRNAs without knowing their exact identification numbers. Under the circumstances that one microRNA query results in multiple IDs, one can further select the microRNAs of the greatest interests. Similarly, Figure 1 provides a snapshot of the query workflow of ‘search by miRNA ID’.
In the miR2Disease system, the concept of target genes can be classified into three categories: the target genes reported in the original reference, the target genes documented in the TarBase system (experimentally validated targets) and the predicted targets from computational tools (Miranda, TargetScan or PicTar). The ‘search’ by ‘target genes’ functionality allows users to search microRNA–disease relationships from the target genes of the first two types (reported targets and experimentally validated targets). Searching through the computationally predicted targets will be included in the future release.
The causal relationship between disease and microRNA is documented and listed in both the searching result page and the detailed relationship page (Figure 1). In addition, the system also allows users to filter the search results by only selecting causal relationships.
Submit page
miR2Disease provides a submission page that allows other researchers to submit important microRNA–disease relationships that are not documented. Once approved by the submission review committee, the submitted record will be included in the database, and made available to the public in the coming release. miR2Disease will be updated monthly.
DISCUSSION
Emerging evidence suggests that specific temporal and spatial microRNA expression is required for normal cellular development and differentiation, whereas many human diseases are associated with aberrant microRNA expression patterns. In order to provide a central resource for the biologists who study the relationship between microRNA and human disease, we develop miR2Disease, a database system aiming at providing a comprehensive resource of microRNA deregulation in various human diseases. miR2Disease not only offers an easy-to-use web interface to query the detailed information on microRNA–disease relationships documented in the database, but it also provides a submission page that allows other researchers to contribute to the data contents.
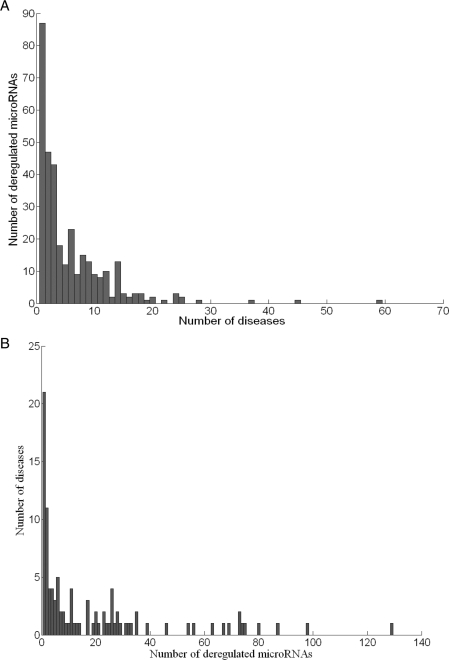
Figure 2A demonstrates the histogram of the number of diseases associated with the microRNAs archived in the database. Eighty-seven microRNAs (29.1%) are deregulated in only one disease; while 177 microRNAs (59.2%) demonstrate deregulation in no more than three diseases. The most prevalent microRNA appeared in the database is hsa-miR-21, which is deregulated in 59 documented diseases (over half of all the disease types). Five additional microRNAs are deregulated in over 25 types of diseases; they are: hsa-miR-155, hsa-miR-221, hsa-let-7a, hsa-miR-223 and hsa-miR-222, which are related to 45, 37, 28, 25 and 25 diseases, respectively. The histogram of the number of deregulated microRNAs associated with individual disease is shown in Figure 2B. No more than three microRNA deregulations were documented for 37 diseases (39.4%), 21 of which contain only one record. On the contrary, 13 diseases are related to more than 50 deregulated microRNAs. Hepatocellular carcinoma, prostate carcinoma and colorectal carcinoma top this list with 129, 98 and 87 deregulated microRNAs associated. One should notice that these numbers are highly dependent on the experimental approaches being used in the original publication. For instance, investigations using microarray technology usually observe more deregulated microRNAs than other low-throughput studies.
Figure 2.
Distribution of diseases and deregulated microRNAs documented in miR2Disease database. (A) Histogram of the number of diseases associated with individual microRNA. (B) Histogram of the number of deregulated microRNAs associated with individual disease.
It remains to be seen whether the deregulation of microRNAs is a cause or consequence of the disease state. Around 1/7 of the microRNA–disease relationships in miR2Disease indicate the pathogenic role of microRNA deregulation in various diseases, including cancer, metabolic disease and cardiovascular disease. For instance, Ma et al. (39) reported that highly expressed miR-10b initiates tumor invasion and metastasis in breast cancer through translational inhibition of HOXD10, and eventually increases expression of RHOC, a pro-metastatic gene. Huang et al. (40) found that significant upregulation of miR-373 and miR-520c stimulates breast cancer cell migration and invasion by the suppression of CD44. Meng et al. (41) identified that the oncogene mitogen-activated protein kinase kinase kinase 8 (MAP3K8) is a target of miR-370, whose reduced expression elevates the MAP3K8 level and therefore contributes to tumor growth in cholangiocarcinoma. It has been reported that miR-375 controls insulin secretion by regulating the expression of myotrophin. Upregulation of miR-375 led to an enhanced inhibition of insulin release (42). It is also demonstrated that downregulation of miR-1 and miR-133 contributes to re-expression of HCN2/HCN4 and the electrical remodeling process in hypertrophic hearts (43).
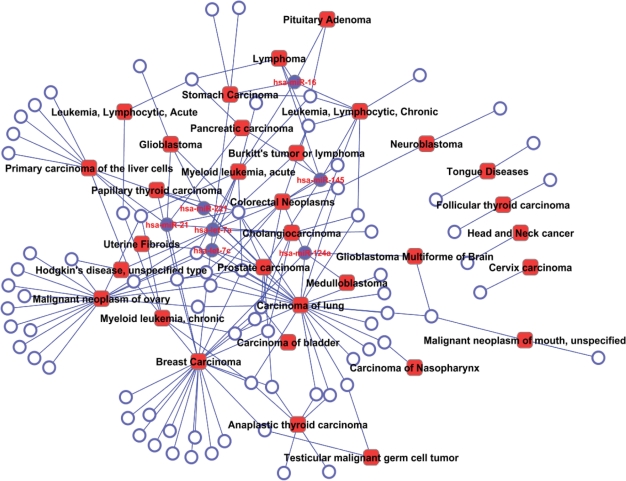
Information in the miR2Disease can be used to describe the relationship among multiple diseases. To this end, we create a bipartite network that describes the causal relationship between 85 microRNAs and 32 cancer-related diseases (Figure 3). Within this network, lung carcinoma, breast cancer and ovarian cancer demonstrate the highest connectivity by affiliating with 25, 23 and 18 causal microRNAs, respectively. For microRNAs, hsa-let-7a deregulation is the causal effect for nine types of cancers, while hsa-miR-21, hsa-miR-124a, hsa-let-7c, hsa-miR-145, hsa-miR-16 and hsa-miR-221, each associate with five types of cancers or more (solid circles in the network). Using miR2Disease database, one can create this type of bipartite network by searching for the diseases or microRNAs of interest.
Figure 3.
A bipartite network demonstrating the causal relationship between deregulated microRNAs and variety of cancers. Circles and rectangles correspond to microRNAs and diseases, respectively. An edge is placed between a microRNA and a disease if the microRNA deregulation plays causative role in pathogenesis of the disease. Solid circles indicate master microRNAs that potentially cause no less than five types of cancers.
miR2Disease documented several potential mechanisms that cause the microRNA deregulation in various human diseases. First, microRNAs are located in the disease-causing genomic loci, including minimal regions of loss of heterozygosity, minimal amplicon, or breakpoint cluster regions (19,25). For instance, miR-15 and miR-16 are located in chromosome 13q14, a region that is deleted in more than half of B-cell chronic lymphocytic leukemia (CLL)s (B-CLL); consequently, both genes are deleted or downregulated in the majority (68%) of CLL cases (44). In contrast, the miR-17-92 polycistron is located in a genomic region that is amplified in human B-cell lymphomas, and therefore are overexpressed (45). Second, microRNA deregulation is caused by abnormal epigenetic patterns, including abnormal DNA methylation and histone-modification patterns (46). For instance, in normal conditions, the putative promoter region of let-7a-3 gene is heavily methylated in normal human tissues, but hypo-methylated in lung adenocarcinoma. Such promoter demethylation induced reactivation of the let-7a-3 with an oncogenic function and increased proliferation in lung cancer cell lines (47). In addition, aberrant hypermethylation leads to miR-9-1 inactivation in human breast cancer (48). Third, microRNA deregulation may be caused by abnormalities of the enzymes that are involved in microRNA biogenesis. For example, Otsuka et al. (49) unambiguously showed that miR-24 and miR-93 could target viral large protein (L protein) and phosphoprotein (P protein) genes. In Dicer1-deficient cells, a lack of host miR-24 and miR-93 was responsible for increased VSV replication. In the miR2Disease system, such information is documented in the ‘analysis’ session (a link through the home page).
In addition to the mechanisms described above, miR2Disease also contains entries where disease-causing genetic variations affect cellular functions through the disruption of microRNA target selection. For instance, loss-of-interaction variations in let-7:Hmga2 (50), miR-148a:HLA-G (51) and miR-433:FGF20 (52), are involved in myoma, asthma and Parkinson's disease, respectively. Chen et al. (53) elucidated that CCND1 mRNA is normally under the regulation of miR-16-1, and that truncated CCND1 mRNA escapes this regulation through deletion of the microRNA-binding sites in mantle cell lymphoma. In summary, loss-of-interaction variations in microRNA genes or the binding site(s) of microRNA represent a newly described pathogenic mechanism of disease.
Overall, miR2Disease provides a comprehensive resource of microRNA deregulation in human disease. We believe that miR2Disease will be of particular interest to the life science community and facilitates the biologists to unravel the role of microRNA in the pathogenesis of human disease.
FUTURE DEVELOPMENT
As stated in the ‘data collection and database content’ session, the microRNA–disease relationships documented in the current release were collected through searching the PubMed database with a list of keywords, such as ‘microRNA disease’, ‘miRNA disease’, ‘microRNA cancer’, ‘miRNA cancer’, etc. Although we collected ∼2000 relationships from over 600 literatures, such mechanism suffers from the lack of comprehensiveness and systematicness. We plan to adopt two strategies to improve our data collection. First, we will use text-mining tools to help us prescreen PubMed abstracts that potentially describe the microRNA–disease relationships. Second, we will specifically target MeSH (Medical Subject Headings) vocabularies that are created and maintained at the National Library of Medicine. These strategies will no doubt increase the data comprehensiveness, and will impact the database records in the releases to come.
DISCLAIMER
Not all relationships of microRNA in disease are equally valid. Some studies demonstrated more creditable microRNA–disease relationships than others.
FUNDING
The China National 863 High-Tech Program (2007AA02Z302 to Y.L. and 2007AA02Z329); the Indiana Genomics Initiative of Indiana University; the Lilly Endowment, Inc. (to Y.L., partial); National Natural Science Foundation of China (Grant No. 60671013). Funding for open access charge: The China National 863 High-Tech Program.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank informative suggestions from Dr Kenneth P. Nephew at Indiana University.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36:D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 16.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 17.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 20.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 21.Nana-Sinkam SP, Geraci MW. MicroRNA in lung cancer. J. Thorac. Oncol. 2006;1:929–931. [PubMed] [Google Scholar]
- 22.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Yang N, Coukos G, Zhang L. MicroRNA epigenetic alterations in human cancer: one step forward in diagnosis and treatment. Int. J. Cancer. 2008;122:963–968. doi: 10.1002/ijc.23325. [DOI] [PubMed] [Google Scholar]
- 24.Blenkiron C, Miska EA. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum. Mol. Genet. 2007;16(Spec No 1):R106–R113. doi: 10.1093/hmg/ddm056. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McManus MT. MicroRNAs and cancer. Semin. Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 28.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 30.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 33.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol. Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 35.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudder A, Novak RF. miRNAs: effectors of environmental influences on gene expression and disease. Toxicol. Sci. 2008;103:228–240. doi: 10.1093/toxsci/kfn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 40.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 41.Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27:378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 42.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 43.Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J. Biol. Chem. 2008;283:20045–20052. doi: 10.1074/jbc.M801035200. [DOI] [PubMed] [Google Scholar]
- 44.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fraga MF, Esteller M. Towards the human cancer epigenome: a first draft of histone modifications. Cell Cycle. 2005;4:1377–1381. doi: 10.4161/cc.4.10.2113. [DOI] [PubMed] [Google Scholar]
- 47.Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 48.Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 49.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 50.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am. J. Hum. Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, Eckhardt SG, Robinson WA. Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood. 2008;112:822–829. doi: 10.1182/blood-2008-03-142182. [DOI] [PMC free article] [PubMed] [Google Scholar]