Abstract
Pseudoknots have been recognized to be an important type of RNA secondary structures responsible for many biological functions. PseudoBase, a widely used database of pseudoknot secondary structures developed at Leiden University, contains over 250 records of pseudoknots obtained in the past 25 years through crystallography, NMR, mutational experiments and sequence comparisons. To promptly address the growing analysis requests of the researchers on RNA structures and bring together information from multiple sources across the Internet to a single platform, we designed and implemented PseudoBase++, an extension of PseudoBase for easy searching, formatting and visualization of pseudoknots. PseudoBase++ (http://pseudobaseplusplus.utep.edu) maps the PseudoBase dataset into a searchable relational database including additional functionalities such as pseudoknot type. PseudoBase++ links each pseudoknot in PseudoBase to the GenBank record of the corresponding nucleotide sequence and allows scientists to automatically visualize RNA secondary structures with PseudoViewer. It also includes the capabilities of fine-grained reference searching and collecting new pseudoknot information.
INTRODUCTION
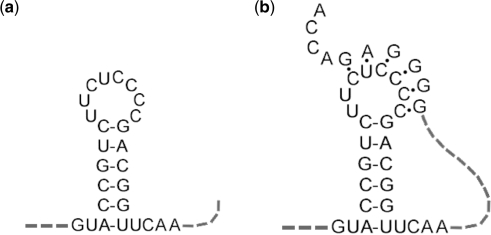
The secondary structure of an RNA molecule is formed by base pairing between various regions of the RNA that results in a configuration of double-helical regions (stems) and single-stranded loops. In general, a RNA pseudoknot is defined as a secondary structure formed by pairing between a loop and a region located outside (upstream or downstream) of the stem flanking the loop. Figure 1a shows a simple stem-loop and Figure 1b shows a simple pseudoknot. Both ‘orthodox’ secondary structures and pseudoknots have been implicated in important biological functions such as gene expression and viral genome replication (1,2). There is an active research community that brings together scientists with diverse expertise to develop and apply computational methods to directly link different types of RNA secondary structures to their biological functions.
Figure 1.
Example of stem-loop (a) and pseudoknot (b).
While pseudoknots have been observed in a variety of eukaryotic and prokaryotic RNA, this type of structures have particularly captured the interests of many virologists because of their frequent involvement in ribosomal frameshift, read-through and other mechanisms of regulation in viral gene expression and replication (3).
PseudoBase (4), available at http://wwwbio.leidenuniv.nl/~Batenburg/PKB.html and and http://www.ekevanbatenburg.nl/PKBASE/, is currently the main public source of information about pseudoknot secondary structures. Yet, to organize, integrate and perform elaborate analysis on specific pieces of information, scientists still have to do a substantial amount of handwork. For example, in order to collect a set of all known pseudoknots which fit certain criteria (e.g. those pseudoknots occurring in viruses that are supported by mutagenesis) and examine them visually, one would have to go through the records in PseudoBase, select those satisfying the criteria and download their records. Afterwards, the nucleotide sequences of retrieved pseudoknots need to be converted to the dot-parentheses format. The sequence and secondary structure data are then submitted to visualization portals such as PseudoViewer (5) in order to view the structures. Ideally, scientists should be able to go through all these steps with a unified, user-friendly interface to PseudoBase that screens them from database issues and provides them with powerful tools for searching, formatting and visualization.
This work extends PseudoBase to PseudoBase++, a searchable, up-to-date database of the PseudoBase pseudoknots wrapped by a versatile, user-friendly interface providing scientists with a powerful engine to access, search, select and sort data based on different fine-grained criteria. The PseudoBase++ interface also allows scientists to visualize selected structures with PseudoViewer, to map existing sequences to GenBank (6), and to insert new pseudoknots to the PseudoBase dataset though a syntax-controlled interface that prevents structural error for long sequences. PseudoBase++ is part of the RNAVLab project—a virtual laboratory for the analysis of RNA secondary structures (7) and serves the specific purpose of facilitating analysis of pseudoknots. Figure 2 shows the PseudoBase++ main Web page.
Figure 2.
The main Web page of PseudoBase++.
REORGANIZATION OF PSEUDOBASE IN A SEARCHABLE DATASET
PseudoBase++ differs in design from the original PseudoBase in the following aspects. First, PseudoBase++ is based on the Model-View-Controller (MVC) architecture, a design in which the domain-specific data (the model of the data) is separated from its presentation (the view of the data). This separation allows for changes in the model without affecting the view and vice versa. It also results in applications that are easier to modify, maintain and extend. Furthermore, the pseudoknot data in PseudoBase++ is stored as a relational database. Relational databases are often used as the model for MVC applications for several reasons such as efficient data storage and retrieval. A relational database for modeling the data in PseudoBase++ also allows for fine-grained searching and filtering capabilities, and facilitates the integration of new functionalities.
To build a relational model out of the PseudoBase dataset, PseudoBase++ uses an engine that polls PseudoBase data periodically (and automatically) so that the model always has the latest information from PseudoBase. Data from the PseudoBase records are extended and inserted into the relational schema. The relational model in PseudoBase++ consists of eight tables (i.e. sequence containing the nucleotide data; structure containing the secondary structure information; pairs, stems, stempairs and loops containing the detailed location of secondary structure motifs and bases bounded; pkrefs pointing to the extensive reference information; and pkreferences containing the detailed reference information). Figure 3 displays the tables, their fields and the field descriptions. The fields mapped from PseudoBase into the PseudoBase ++ tables are: PseudoBase identifier (PKB number), definition, organism, abbreviation, RNA type, keywords, EMBL number, submitted by, supported by, stem size, loop size, positions paired, start, end, sequence, brackets, length and references. Additionally, new fields include the classification of pseudoknots per type (pseudoknot type) as presented in ref. (5) and a fine-grained reference dataset based on BibTeX fields. PseudoBase++ allows scientists to easily submit new pseudoknots. Thus, a twin schema, separated from the schema containing the PseudoBase dataset, is used to capture user-submitted data.
Figure 3.
Diagram for the PseudoBase++ database table relationships.
INTEGRATION OF NEW FUNCTIONALITIES
Design and implementation of new functionalities
PseudoBase++ is implemented using Ruby on Rails 2.1 and MySQL running via Apache on the OpenSuse Linux platform. Ruby on Rails (or Rails for short) is an open-source framework for Web applications using the Ruby object oriented scripting language. Advantages of using Rails for building front-ends include fast development, easy maintenance and code reusability. Ruby is a high level interpreted language that is very easy to learn and use. The Rails framework includes out of the box scripts that perform common tasks including database migration, automatic link routing, unit testing and generation of application components. The basic framework is powerful, extensible, as well as easy to learn and use, making maintenance simple. Code written within the framework is highly reusable because it stresses ‘convention over configuration’. Rails use the familiar MVC design pattern to streamline applications into manageable and interdependent pieces, which can be modified to suit an the application's specific needs. An entirely new application could be built directly over PseudoBase++ by changing the outward appearance and database schema.
The interface to the PseudoBase++ functionalities has been designed and implemented with the scientist's requirements in mind and therefore it can be defined as scientist-centric. Effective access to information includes fast, easy to use functionalities as well as flexibility in defining the scientist's requirements. The scientist should be able to quickly search across all the database fields by using a one-line text box for quick searching, much like in familiar search engines such as Google and e-Bay. At the same time the interface should include advanced and refined search opportunities in which the scientist can define a more detailed search either after the initial quick search or from the beginning by clicking on the advanced search link on the main page. The scientist expects to be able to toggle what fields in the page should be displayed and the result page should contain all the pertinent information for a sequence according to the scientist's preferences. PseudoBase++ should also serve as a learning environment for beginners with simple information about pseudoknots and RNA in general.
Guided by the features above, we included these new functionalities in PseudoBase++:
Simple and advanced search across PseudoBase database based on keywords.
Fine-grained search of bibliography keywords, e.g. author name, journal and year.
Data formatting (i.e. FASTA, dot-parentheses and BPSEQ formats).
Mapping to GenBank and visualization with PseudoViewer.
Classification of pseudoknots per type (pseudoknot type).
Submission of new pseudoknots.
Deploying of Web services to access the database.
Simple and advanced search across PseudoBase database based on keywords
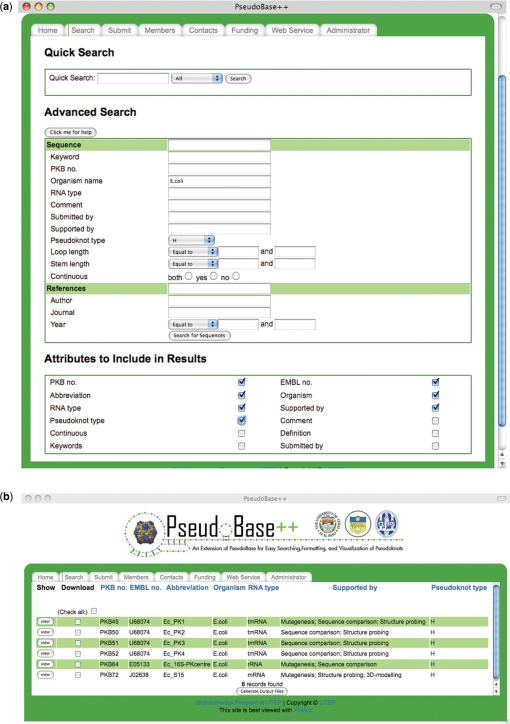
The front-end interface allows users to query the database based on any keyword in any field of the database (simple search) or based on a number of search parameters such as organism, abbreviation, sequence length, RNA type and reference (advanced search). The search can accommodate combinations of search parameters and the user can choose the parameters to visualize in the final list of results among the fields of the database. After the search is submitted, the list of the pseudoknots in the database satisfying the specified criteria is returned. Figure 4 shows an example of search with its input parameters, i.e. Organism = E.coli AND pseudoknot type = H, in Figure 4a and its output results in Figure 4b.
Figure 4.
Example of search (a) browser with input parameters and (b) browser with results for the given input parameters.
Fine-grained bibliography search
In many situations, important information about a pseudoknot needs to be retrieved from related published work. Each pseudoknot record in the original PseudoBase contains a reference list. In PseudoBase++, we store citations in the BibTeX format a detailed description of which can be found at http://www.bibtex.org/Format. This opens up the possibility to conduct fine-grained reference searches. We provide the functionality of searching by author name, journal title and publication year in the current version of PseudoBase++ and will extend to other useful features in the future.
Data formatting
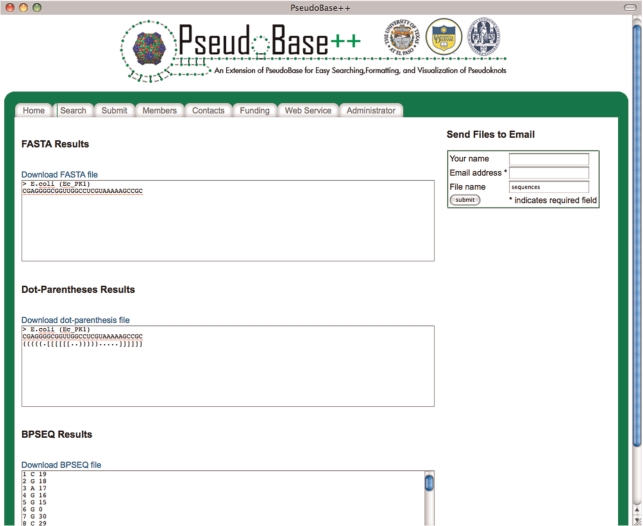
Data related to nucleotide sequences can be extracted in FASTA, dot-parentheses and BPSEQ formats. The information can be downloaded in a file that is available to the user for 7 days after the request has been submitted or that is sent to a recipient though an e-mail. FASTA files can ultimately be used as input for secondary structure prediction algorithms such as PKnots-RE (8), PKnots-RG (9) and NuPack (10). Submissions to any of these codes can be done with RNAVLab (7). Figure 5 shows an example of the three formats for a pseudoknot in PseudoBase++.
Figure 5.
Example of the three formats for a pseudoknot in PseudoBase++.
Mapping to GenBank and visualization with PseudoViewer
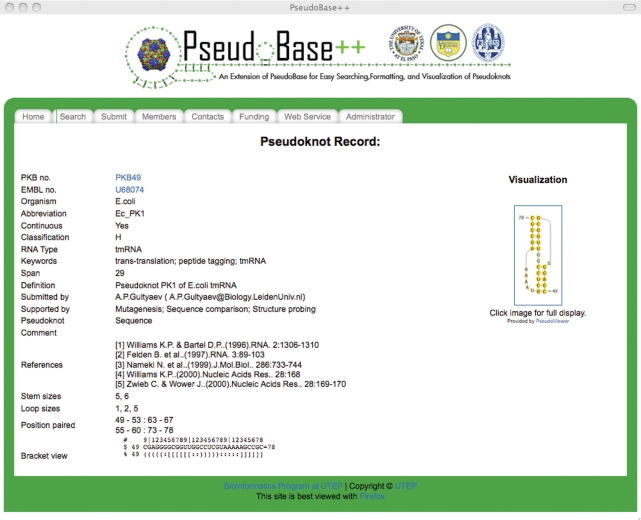
The entries returned from a search are numbered sequentially. By clicking on View, the user can access a pseudoknot description page or pseudoknot record containing detailed information about the pseudoknot, including its secondary structure visualization (Figure 6). The accession number (or EMBL no.) in the pseudoknot record allows the user to access the GenBank information for the selected sequence while the PKB number (or PKB no.) allows the user to access the PseudoBase information.
Figure 6.
Pseudoknot record of the PKB49 pseudoknot in PseudoBase++.
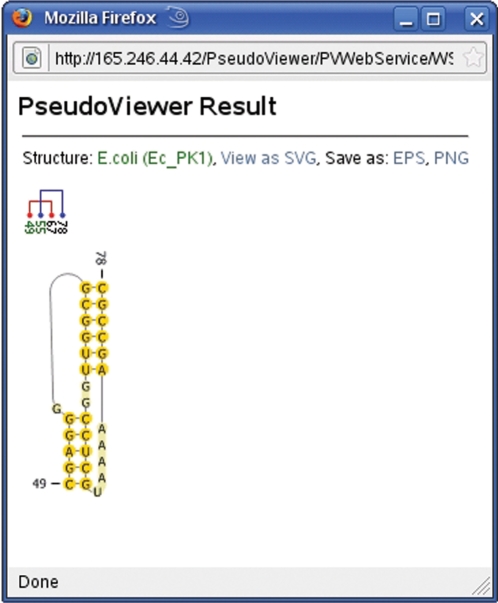
By clicking on the visualization of the pseudoknot in the pseudoknot record, the user invokes the ‘on the fly’ visualization feature (Figure 7). The nucleotide sequence as well as the base pairing in the secondary structure are transferred to the PseudoViewer Web page using HTML forms, generating a visualization of the pseudoknot. The visualized pseudoknot can be saved in EPS and PNG formats.
Figure 7.
Example of ‘on the fly’ visualization of a pseudoknot with PseudoViewer.
Classification of pseudoknot type
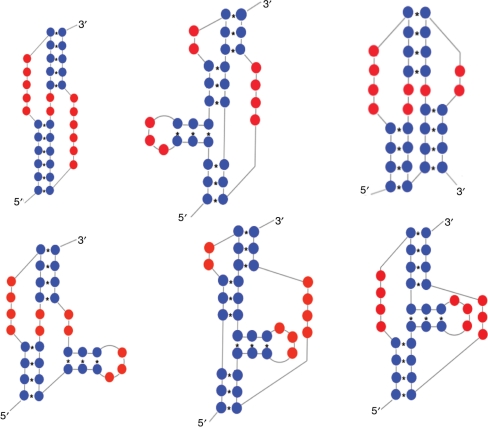
The classification of pseudoknots per type (pseudoknot type) provided in PseudoBase++ is based on Han and Byun (5) and clusters the secondary structures into six different simple types, i.e. H-, HH-, HHH-, HL_out-, HL_in- and LL_in-type as well as an unknown or unclassified type. Note that ‘H’ means hairpin loop, ‘L’ means bulge loop, ‘in’ means internal loop or multiple internal loops, and ‘out’ means external loop or multiple external loops. The classification provided by our tool works on the string of brackets to extract the proper type that is provided for each sequence in the interface. Figure 8 shows the six pseudoknot types.
Figure 8.
Pseudoknot types, i.e. H-, HH-, HHH-, HL_out-, HL_in- and LL_in-type.
Submission of new pseudoknots
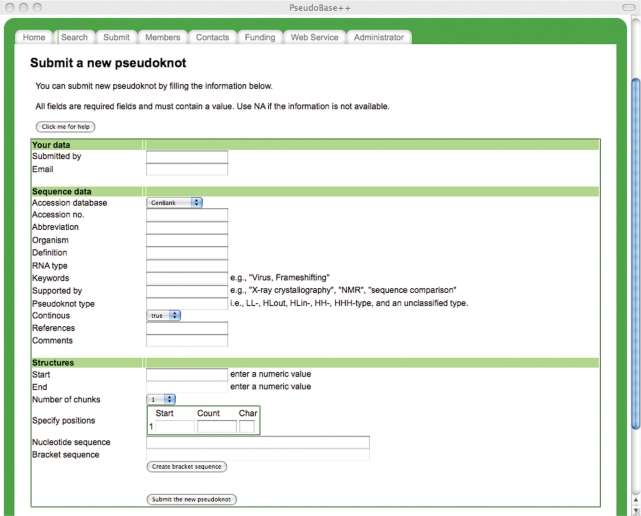
The Submit functionality allows users to easily send us information for a new pseudoknot (Figure 9). To reduce the chance for input errors, pseudoknots are entered by bracket or colon chunks depicting the sequence folding. Users only need to enter the number of chunks, their lengths and the character type, i.e. brackets or colons. The loop size, stem size and paired positions are calculated automatically and the checking for consistencies is performed. Users can verify the structure by submitting the temporary sequence to PseudoViewer that generates a structure only if the base pairing is valid. Newly sent pseudoknots are validated before being added to the PseudoBase and mapped in PseudoBase++. The validation and integration of new structures in PseudoBase are done by Van Batenburg and Gultyaev at the University of Leiden (The Netherlands).
Figure 9.
Web page for the submission of new pseudoknot in PseudoBase++.
Deployment of Web services to access the database
PseudoBase++ provides a simple Web service that allows users to write client applications to access our database directly. Given a simple search query, our service returns all the pseudoknot records matching the query. Our Web service is described using the Web Services Description Language (WSDL) and can be accessed via SOAP and XMLRPC. Many languages such as Java, Ruby, PHP, C# have APIs for this protocol and can automatically generate ready-made clients given the WSDL file.
SUMMARY
In this article, we present PseudoBase++, an extension of the PseudoBase database containing RNA pseudoknots that includes new functionalities to facilitate access to pseudoknot information on a single platform. Among the several functionalities presented in this article, PseudoBase++ allows for efficient compound searches for general and advanced results, extracts sequences in FASTA, dot-parentheses and BPSEQ formats to be used for secondary structure prediction algorithms, automates visualization of structures through PseudoViewer and integrates robust and user-friendly mechanism for adding new pseudoknots to the database. The framework for PseudoBase++ can be easily adapted and used for other categories of RNA and DNA databases.
FUNDING
Texas Advanced Research Program (003661-0008-2006, 003661-0013-2007); the National Institutes of Health (S06GM08012-38, 5G12RR008124-11, 3T34GM008048-20S1); the National Science Foundation (DMS0800272), and the CRA-W Distributed Research Experiences for Undergraduates Program 2008. Funding for open access charge: National Institutes of Health (S06GM08012-38).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
The authors thank Mr Yash Dayal for his assistance in implementing an earlier version of this database.
REFERENCES
- 1.Jones-Rhoades W, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 2.Lyubetsky VA, Pirogov SA, Rubanov LI, Seliverstov AV. Modeling classic attenuation regulation of gene expression in bacteria. J. Bioinform. Comput. Biol. 2007;5:155–180. doi: 10.1142/s0219720007002576. [DOI] [PubMed] [Google Scholar]
- 3.Brierley I, Pennell S, Gilbert RJC. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Batenburg FHD, Gultyaev AP, Pleij CWA, Ng J, Oliehoek J. PseudoBase: a database with RNA pseudoknots. Nucleic Acids Res. 2000;28:201–204. doi: 10.1093/nar/28.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han K, Byun Y. PseudoViewer2: visualization of RNA pseudoknots of any type. Nucleic Acids Res. 2003;31:3432–3440. doi: 10.1093/nar/gkg539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taufer M, Leung MY, Solorio T, Licon A, Mireles D, Johnson KL. RNAVLab RNAVLab: a virtual laboratory for studying RNA secondary structures based on grid computing technology. J. Parallel Comput. 2008;34:661–680. doi: 10.1016/j.parco.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivas E, Eddy SR. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 1999;285:2053–2068. doi: 10.1006/jmbi.1998.2436. [DOI] [PubMed] [Google Scholar]
- 9.Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007;49:65–88. [Google Scholar]