Abstract
ConsensusPathDB is a database system for the integration of human functional interactions. Current knowledge of these interactions is dispersed in more than 200 databases, each having a specific focus and data format. ConsensusPathDB currently integrates the content of 12 different interaction databases with heterogeneous foci comprising a total of 26 133 distinct physical entities and 74 289 distinct functional interactions (protein–protein interactions, biochemical reactions, gene regulatory interactions), and covering 1738 pathways. We describe the database schema and the methods used for data integration. Furthermore, we describe the functionality of the ConsensusPathDB web interface, where users can search and visualize interaction networks, upload, modify and expand networks in BioPAX, SBML or PSI-MI format, or carry out over-representation analysis with uploaded identifier lists with respect to substructures derived from the integrated interaction network. The ConsensusPathDB database is available at: http://cpdb.molgen.mpg.de
INTRODUCTION
Functional interactions between cellular entities like genes, proteins, metabolites, etc. are the key drivers of cellular functions. Different experimental methods like chromatin immunoprecipitation (1) and two-hybrid assays (2), among others, have generated large amounts of interaction data for many organisms, usually stored in interaction databases. In the past few years, the analysis of interaction networks has become crucial to understand biological processes and their dysfunctions in human diseases. For example, reaction networks build the basis of computational models in systems biology. Analyses combining expression and interaction data have recently been used to reveal previously unknown disease mechanisms (3,4). Thus, collecting comprehensive human interaction data is the key to gain new insights into cell biology.
While for several model organisms like Saccharomyces cerevisiae (5) and Caenorhabditis elegans (6), such comprehensive functional interaction networks are available, the larger part of the human interactome remains undiscovered (7). Even worse, the existing knowledge on human functional interactions is dispersed in over 200 interaction databases, each of which has a specific data format, focus and bias (8). Most integration efforts with respect to interaction data so far have focused on merging homogeneous interaction networks. For example, APID (9), MiMI (10) and UniHI (11) integrate protein–protein interaction networks from multiple sources. However, the integration of heterogeneous interactions remains a challenge. Such integration is highly relevant because the resulting network reflects multiple functional aspects of the nodes at the same time (like regulatory relations, physical interactions, catalyzed reactions), and thus constitutes a more complete picture of the living system.
We have developed ConsensusPathDB, a database for integrating human molecular interaction networks, in order to address such a comprehensive integration of interaction data. The integrated content comprises different types of functional interactions that interconnect diverse types of cellular entities. In order to gain an immediate critical number of interactions, we have focused primarily on the integration of existing database resources although our schema has also been used for additional manual upload of experimental interactions. Currently, the database contains human functional interactions, including gene regulations, physical (protein–protein and protein-compound) interactions and biochemical (signaling and metabolic) reactions, obtained by integrating such data from 12 publicly accessible databases (referred to as source databases): Reactome (12), KEGG (13) (metabolic reactions only), HumanCyc (14), PID (http://pid.nci.nih.gov), BioCarta (http://www.biocarta.com), NetPath (http://www.netpath.org), IntAct (15) (data from small-scale experiments only), DIP (16), MINT (17), HPRD (18), BioGRID (19) and SPIKE (20). In this article, we describe the methods used for data integration, the database schema, as well as the main functions of the web interface.
RESULTS
Mapping of functional interactions
In order to assess the content overlap of the source databases and to reduce redundancy, we have applied a method to merge identical physical entities and identify similar interactions. The method is straightforward and efficient for the integration of networks from any single species. Simple physical entities of the same type (genes, proteins, transcripts, metabolites) are compared on the basis of common database identifiers like UniProt (21), Ensembl (22), Entrez (23), ChEBI (24), etc. Since different databases tend to annotate physical entities with different identifier types (e.g. some databases annotate proteins with UniProt identifiers, others with Ensembl identifiers), we first translated the annotations to a uniform identifier type, which is a UniProt entry name in case of proteins, Ensembl gene ID in case of genes and transcripts, and KEGG/ChEBI ID in case of metabolites. Protein complexes are compared according to their individual protein composition. Simple physical entities with the same identifier, and complexes with the same composition, are merged in ConsensusPathDB. Information provided by the according source databases for the merged entities is stored in a complementary manner.
Functional interactions of physical entities are also compared with each other. Here, we distinguish between primary and secondary interaction participants. Primary participants are substrates and products in case of biochemical reactions, interactors in case of physical interactions and target genes in case of gene regulation. All other participants, e.g. enzymes and interaction modifiers, are secondary participants. If the primary participants of two or more interactions match, these interactions are considered similar. Two similar interactions may have different stoichiometry, modification and/or localization of the participants. To allow for flexibility, similar interactions are marked as such in the database, but the decision whether they should be considered identical despite mismatching details is left to the user and depends on his specific problem. Moreover, ConsensusPathDB does not provide any additional quality control filters. All interactions provided by the different database sources are treated in the same way. The results of our mapping method applied on the data from the source databases mentioned above are summarized in Table 1.
Table 1.
Database content and pairwise database overlaps in terms of matching physical entities and similar interactions
| Reactome | Kegg | Humancyc | Pid | Biocarta | Netpath | Intact | Dip | Mint | Hprd | Biogrid | Spike | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Database overlaps in terms of matching physical entities | ||||||||||||
| Reactome | 6831 | 1037 | 893 | 560 | 796 | 241 | 1067 | 352 | 1217 | 2041 | 1704 | 1506 |
| Kegg | 1037 | 3271 | 1482 | 59 | 383 | 15 | 225 | 23 | 329 | 1297 | 540 | 508 |
| Humancyc | 893 | 1482 | 3892 | 253 | 688 | 110 | 553 | 138 | 701 | 1656 | 1065 | 947 |
| Pid | 560 | 59 | 253 | 3614 | 764 | 354 | 799 | 371 | 886 | 1357 | 1263 | 1027 |
| Biocarta | 796 | 383 | 688 | 764 | 3387 | 302 | 892 | 392 | 1011 | 1695 | 1477 | 1346 |
| Netpath | 241 | 15 | 110 | 354 | 302 | 739 | 392 | 173 | 450 | 615 | 568 | 479 |
| Intact | 1067 | 225 | 553 | 799 | 892 | 392 | 4138 | 576 | 2660 | 3546 | 3016 | 3237 |
| Dip | 352 | 23 | 138 | 371 | 392 | 173 | 576 | 964 | 637 | 865 | 828 | 707 |
| Mint | 1217 | 329 | 701 | 886 | 1011 | 450 | 2660 | 637 | 5849 | 4684 | 3939 | 3915 |
| Hprd | 2041 | 1297 | 1656 | 1357 | 1695 | 615 | 3546 | 865 | 4684 | 10165 | 7185 | 5973 |
| Biogrid | 1704 | 540 | 1065 | 1263 | 1477 | 568 | 3016 | 828 | 3939 | 7185 | 8696 | 5248 |
| Spike | 1506 | 508 | 947 | 1027 | 1346 | 479 | 3237 | 707 | 3915 | 5973 | 5248 | 7012 |
| Database overlaps in terms of similar interactions | ||||||||||||
| Reactome | 4129 | 262 | 122 | 101 | 80 | 34 | 97 | 31 | 51 | 304 | 212 | 118 |
| Kegg | 262 | 1655 | 213 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Humancyc | 122 | 213 | 1322 | 0 | 2 | 0 | 1 | 2 | 2 | 7 | 4 | 2 |
| Pid | 101 | 0 | 0 | 3510 | 264 | 96 | 63 | 46 | 73 | 333 | 243 | 186 |
| Biocarta | 80 | 4 | 2 | 264 | 2221 | 67 | 51 | 35 | 43 | 139 | 114 | 173 |
| Netpath | 34 | 0 | 0 | 96 | 67 | 1915 | 57 | 34 | 123 | 821 | 510 | 224 |
| Intact | 97 | 0 | 1 | 63 | 51 | 57 | 6880 | 312 | 2714 | 3216 | 1583 | 4080 |
| Dip | 31 | 0 | 2 | 46 | 35 | 34 | 312 | 1218 | 389 | 821 | 656 | 418 |
| Mint | 51 | 0 | 2 | 73 | 43 | 123 | 2714 | 389 | 13187 | 7211 | 4428 | 5676 |
| Hprd | 304 | 0 | 7 | 333 | 139 | 821 | 3216 | 821 | 7211 | 37955 | 19463 | 11351 |
| Biogrid | 212 | 0 | 4 | 243 | 114 | 510 | 1583 | 656 | 4428 | 19463 | 28038 | 10303 |
| Spike | 118 | 0 | 2 | 186 | 173 | 224 | 4080 | 418 | 5676 | 11351 | 10303 | 22232 |
Current versions of the integrated databases are: Reactome 25, KEGG 47.0, HumanCyc 12.1, PID 2008_06_10, BioCarta 2008_01_08, NetPath downloaded on 6.7.2008, IntAct 2008-06-27, DIP 2008-01-13, MINT 2008-05-19, HPRD I_090107, BioGRID 2.0.42 and SPIKE downloaded on 6.7.2008.
Biological pathways in ConsensusPathDB are represented as sets of interactions, whose compositions are adopted from the source databases. This means that individual interactions rather than entire pathways from different databases are compared with each other. This was necessary because the concept of pathway is defined very differently in the respective source databases and the pathway boundaries are rather unclear. For example, KEGG's Glycolysis/gluconeogenesis pathway contains 31 reactions whereas Reactome's Glycolysis contains 10 reactions.
Database schema and content
Interaction data in ConsensusPathDB currently originates from 12 interaction databases and comprises physical interactions, biochemical reactions and gene regulations. The data are gathered in different formats: database dumps, standard interaction file formats like BioPAX (25) or PSI-MI (26), database-specific flat or XML files, or is retrieved through web services. All interaction data are translated to the schema of our relational database and are integrated using the method described above. The database has a graph-like architecture: its three main classes are ‘Interaction’, ‘Physical entity’ and ‘Edge’. The first two classes store information about different interaction types and physical entities, respectively, and edges connect both, indicating that a specific entity participates in a specific interaction. Edges are carriers of information on the particular role, stoichiometry, state and compartment of the physical entity in the particular interaction, if such information is available from the source databases. Several other classes in our database contain information about the source databases, external identifiers of entities or interactions, literature references, cellular compartments, biological pathways, etc. Importantly, the source of physical entities and interactions is always recorded, which allows linking to the original data in the source database.
The database content is updated automatically on a regular basis with the latest releases of the source databases.
Web interface
ConsensusPathDB is accessible via the Internet at http://cpdb.molgen.mpg.de. The main functions of the web interface are described here and summarized graphically in Figure 1. The web interface gives information about the database and its current content summarized in overlap tables (Table 1). It is obvious that the content of existing human interaction resources is partially overlapping but, more importantly, complementary to a large extent. The web interface contains documentation in form of a regularly updated tutorial intended to guide the user through the different functions of the web interface and provide further details (Supplementary Material 1).
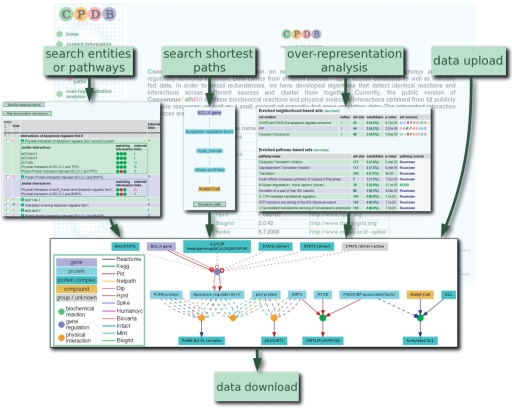
Figure 1.
Graphical summary of the main functions of the ConsensusPathDB web interface. The search for interactions of specific entities or pathways, search for interaction paths, overrepresentation analysis and model upload induce specific interaction subnetworks that can be visualized in the dynamic visualization environment and exported in BioPAX format for further computational analysis, or as images. Different colors of nodes and edges refer to different types of interactions or entities and different source databases, respectively. The background image shows a screenshot of the ConsensusPathDB start page.
Search functions
The user can search for interactions of specific physical entities or pathways by name or database identifiers. Rules according to which interactions with the same primary participants but different compartment/modification/stoichiometry information are to be merged can be specified here. Interactions of interest are merged with their similar counterparts according to these rules (Figure 2), and are displayed as network graphs in the visualization environment of the ConsensusPathDB web interface. In these graphs, two classes of nodes exist: physical entity nodes and interaction event nodes. Node colors encode the specific object type (protein, metabolite, etc.; physical interaction, biochemical reaction, etc.). Edges connect interactions with physical entities and indicate which physical entities participate in which interactions. Different edge styles encode the roles of the entities in the interactions, and edge colors refer to the source of this annotation. The network graphs are automatically generated and dynamical. For example, interactions can be removed from the graph, or new ones can be added by expanding a specific physical entity. Details about nodes, like alternative names and external identifiers, are shown in tool-tips. Physical entities can be easily located in large graphs by searching them by name. Network graphs can be exported as image or as a computer-readable file (currently, in BioPAX level 2 format). In the latter case, networks extracted from ConsensusPathDB can be used as input to various software programs for further analysis, e.g. for modeling and simulation studies.
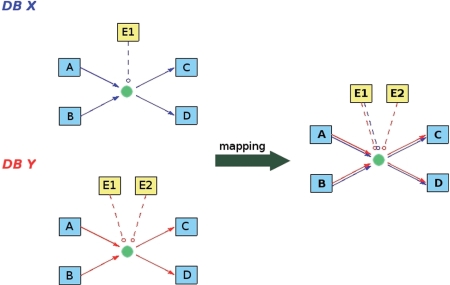
Figure 2.
Illustration of the mapping procedure. Two biochemical reactions that are identical according to their primary participants and the user-specified mapping criteria are mapped. The reaction A + B -> C + D, catalyzed by enzyme E1, originates from database X. A similar reaction, catalyzed by enzymes E1 and E2, originates from database Y. Since both reactions are identical according to the user-specified rules, the interactions are merged and visualized reflecting the source annotations.
Apart from the search for interactions of single physical entities, the user can search for shortest paths of interactions connecting two distinct physical entities in the overall interaction network stored in the database. If a path between the entities of interest exists, it can be further constrained by forbidding certain intermediates. Interaction paths can be visualized in the visualization environment.
Overrepresentation analysis
Using the web interface, over-representation analysis can be carried out with gene sets and functional modules derived from two methods. The first method incorporates pathway definitions as given by the source databases. The second method is based on functional modules defined by proximity measures using the integrated network structure. It includes a node (for example a gene) as the module centre and its neighbours within a user-specified interaction proximity radius. For example, modules with radius 1 include the central gene and all its direct neighbors (genes that appear in a functional interaction together with the central one), and modules with radius 2 additionally include the neighbours of the neighbours of the central gene. Moreover, interconnectivity of module members can be specified by the user with the clustering index. For each predefined module, a P-value is calculated based on the hypergeometric distribution. The P-value reflects the significance of the observed overlap between the input gene list and the module's members as compared to random expectations. A small P-value indicates that more of the module's members are present in the input list than expected by chance. If, for example, the input list contains differentially expressed genes from a case–control study, overrepresentation analysis may point to pathways and functional sub-networks that are dysregulated in the disease state. Overrepresented modules are shown in downloadable lists sorted by the significance of over-representation (P-value) and modules of interest can be visualized in order to see the specific relations between their members.
Import, export and expansion of networks
Apart from the possibility to export interaction networks from the visualization environment, the user can upload an interaction network file in any of three different formats: BioPAX (level 2), PSI-MI (level 2.5) and SBML (level 2) (27). If the physical entities from the uploaded file are annotated with external identifiers, these entities and their interactions are mapped to the interaction network in the database and matching information is indicated. Thereby, the model can be validated against existing interaction knowledge stored in the database, and extended in the context of the database content.
DISCUSSION
ConsensusPathDB is a database that integrates interaction data from heterogeneous resources for human functional interactions in order to create a more complete and less biased picture of the cellular interactions. It can be used in many ways—for example, to retrieve network topologies necessary for mathematical simulations, to interpret gene lists, to assess the distribution of interaction knowledge across interaction databases, or to carry out topological analysis with the integrated human interaction network, which, according to our analysis, provides quite different results compared to topological analysis of the separate interaction databases (results will be published elsewhere).
Although we apply our best efforts to collect and integrate available interaction data, more data sources remain to be integrated. Importantly, human gene regulatory interactions are currently weakly represented in our database (283 interactions) because on the one hand, such data are rare compared to other interaction types (e.g. protein–protein interactions), and, on the other hand, access to the majority of existing gene regulatory data is mostly limited by license constraints (28, 29). Apart from physical interactions, biochemical reactions and gene regulatory interactions, other relations exist between cellular entities that will be integrated into the database, for example, genetic and epigenetic relationships or relations with respect to experimental co-regulation patterns or to more general co-occurrences. Some of these relations are present e.g. in the STRING database (30) and can be integrated into ConsensusPathDB. Although ConsensusPathDB is currently focused on Homo sapiens, the integration of data from other species is an ongoing issue since it will reveal conserved and species-specific cellular processes on the interaction level.
Interaction maps are of great importance in many areas of life sciences, for example in systems biology and molecular medicine. However, more work remains to be done in order to assemble a complete map of the human functional interactome. ConsensusPathDB marks a first step towards achieving this goal by collecting, integrating and interpreting heterogeneous interaction knowledge.
AVAILABILITY
ConsensusPathDB is available freely to academic users via http://cpdb.molgen.mpg.de. Data in form of flat files are available upon request (please contact kamburov@molgen.mpg.de).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the EMBRACE and CARCINOGENOMICS projects that are funded by the European Commission within its 6th Framework Programme under the thematic area ‘Life Sciences, Genomics and Biotechnology for Health’ (LSHG-CT-2004-512092 and LSHB-CT-2006-037712); 7th Framework Programme project APO-SYS (HEALTH-F4-2007-200767); German Federal Ministry of Education and Research within the NGFN-2 program (SMP-Protein, FKZ01GR0472); Max Planck Society within its International Research School program (IMPRS-CBSC). Funding for open access charge: European Commission.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to the interaction database providers (Table 1) that allowed automated access to their databases. Integration of interaction data could only be achieved because the original data was provided in an excellently documented way.
REFERENCES
- 1.Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front. Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]
- 2.Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 3.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maraziotis IA, Dimitrakopoulou K, Bezerianos A. Growing functional modules from a seed protein via integration of protein interaction and gene expression data. BMC Bioinformatics. 2007;8:408. doi: 10.1186/1471-2105-8-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart GT, Ramani AK, Marcotteha EM. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prieto C, De Las Rivas J. APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayapandian M, Chapman A, Tarcea VG, Yu C, Elkiss A, Ianni A, Liu B, Nandi A, Santos C, Andrews P, et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res. 2007;35:D566–D571. doi: 10.1093/nar/gkl859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaurasia G, Iqbal Y, Hänig C, Herzel H, Wanker EE, Futschik ME. UniHI: an entry gate to the human protein interactome. Nucleic Acids Res. 2007;35:D590–D594. doi: 10.1093/nar/gkl817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct – open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatr-Aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database – 2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkon R, Vesterman R, Amit N, Ulitsky I, Zohar I, Weisz M, Mass G, Orlev N, Sternberg G, Blekhman R, et al. SPIKE – a database, visualization and analysis tool of cellular signaling pathways. BMC Bioinformatics. 2008;9:110. doi: 10.1186/1471-2105-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luciano JS. PAX of mind for pathway researchers. Drug Discov. Today. 2005;10:937–942. doi: 10.1016/S1359-6446(05)03501-4. [DOI] [PubMed] [Google Scholar]
- 26.Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, et al. The HUPO PSI's molecular interaction format – a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 27.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 28.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang C, Xuan Z, Zhao F, Zhang MQ. TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 2007;35:D137–D140. doi: 10.1093/nar/gkl1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. STRING 7 – recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]