Abstract
Genome wide scans have shown that positive selection is relatively frequent at the molecular level. It is of special interest to identify which protein sites and which phylogenetic branches are affected. We present Selectome, a database which provides the results of a rigorous branch-site specific likelihood test for positive selection. The Web interface presents test results mapped both onto phylogenetic trees and onto protein alignments. It allows rapid access to results by keyword, gene name, or taxonomy based queries. Selectome is freely available at http://bioinfo.unil.ch/selectome/.
INTRODUCTION
Recent analyses of genomic data have shown that positive selection was more widespread at the molecular level than expected under a restrictive interpretation of the neutral theory (1,2). Positive selection is expected to result in changes in function, and better adaptation of the organism to its environment. As such, identifying which genes are affected by positive selection is an important step not only in studies of evolutionary biology, but also in functional studies (e.g. site-directed mutagenesis).
Recent progress in methods allows to specifically detect positive selection affecting only a few sites on a single branch of the phylogeny (3–5). But such methods are available in computer packages which can be difficult to use for most biologists [e.g. Phylogenetic Analysis by Maximum Likelihood (PAML), 6], and the application to large scans poses problems both of computational power and of test repetition. The latter issue can be solved by the careful use of statistical corrections (2,7). The issue of computational power is due to the need to compute two likelihood values for each branch tested and each gene family studied.
We present a database, Selectome, which includes the results of systematic branch-site tests for positive selection (8) on all internal branches of selected clades, based on gene families from the database TreeFam (9,10). Computations are done for each release of TreeFam, followed by correction for multiple testing over the whole dataset. Results are presented in a Web interface which is voluntarily close to that of TreeFam, to provide users with a similar experience. Selectome is freely available at http://bioinfo.unil.ch/selectome/.
DATA AND METHODS
All definitions of gene families, sequence alignments and phylogenetic trees, come from TreeFam (9,10). We use the ‘clean trees’ of TreeFam, which have been confirmed by several methods. Selectome also includes other tables from TreeFam (10), notably cross-references to Ensembl identifiers.
We do not apply tests for positive selection to all branches of these phylogenies, which span all animal evolution, plus in some cases yeast and plant outgroups. It has not been shown that codon models of positive selection can be robustly used for very ancient branches, such as the split between protostomes and deuterostomes in animals, so we only test more recent subtrees. Moreover, the test used has low power when few sequences are used (2,7), so we do not analyze subtrees with a low number of species, nor do we test terminal branches. In TreeFam release 4 (Selectome release 1), only one subtree fulfils these conditions, the vertebrate clade. For each subtree, the corresponding DNA alignment is extracted, and a well aligned subset of sites is selected using GBLOCKS (11) (type = codons; minimum length of a block = 4; no gaps allowed). It should be noted that despite our best efforts, alignment errors can be a cause of spurious detection of positive selection. This is especially a concern on branches leading to a few closely related species, where gene prediction errors can be propagated between genomes. We invite users to always consider the alignment (see Web interface) before concluding on the presence of positive selection.
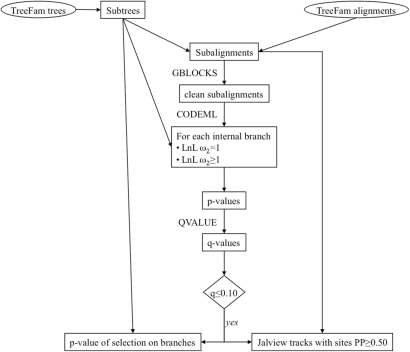
Methods used in Selectome follow closely those of Studer et al. (2) (Figure 1). Briefly, we apply the branch-site model A (8,12) to each internal branch of each subtree tested, as implemented in CODEML from the PAML package version 4b (6). Positive selection is detected if there is a category of sites with dN/dS ratio ω > 1 on the tested branch. Importantly, the test contrasts positive selection on the branch of interest to the possibility of relaxed purifying selection on that branch, which avoids a major source of false positive results (13). The test is done by comparing the difference of log-likelihood values 2 × ΔlnL to a χ2 distribution of 1 degree of freedom. Thus for each branch, the likelihood of the data is estimated twice, under the model which allows positive selection, and under the model which does not. For a small gene family of 14 sequences, which has 11 internal branches in the (unrooted) phylogeny, 22 likelihood values must be estimated. We control for false discovery by using the q-value (14), over all P-values treated as one series of repetitions (m branches × n trees). We use the ‘bootstrap’ method for estimating π0 in the R package QVALUE (Dabney,A. and Storey,J.D., unpublished data), because the P-value distribution is bimodal (2). Selectome reports branches as significant with a threshold of q = 10% of false positives. This may correspond to a different P-value from one release to the next. For branches which pass the threshold, the P-value of the likelihood test is added to the tree file in NHX format, which already contains annotations from TreeFam. These notably include taxonomy, as well as the speciation or duplication type of the branch, inferred by the Duplication/Loss Inference algorithm (9,10).
Figure 1.
Flowchart of methods used to annotate positive selection in Selectome.
When positive selection is significant for a gene on a branch, specific sites under positive selection are detected by Bayes empirical Bayes (15). Selectome reports all sites with a posterior probability (PP) ⩾50%, color coded according to PP value.
Selectome release 1, corresponding to TreeFam-A release 4, contains 1031 subtrees, with 13 062 branches tested and 867 found to contain positive selection. At time of submission, computations are under way for Selectome release 2, corresponding to TreeFam release 6. Selectome is stored in a MySQL database. A complete download is available on the website.
WEB INTERFACE
The Web interface of Selectome provides a user experience which is voluntarily close to that of TreeFam, to facilitate navigation between both resources. Selectome can be queried by a ‘Basic search’ or by one of two advanced query tools. The basic search is a query on all names and annotations of genes and gene families. Thus a query for ‘kinase’ will return 575 genes and 53 gene families (in release 1).
The first type of advanced query is similar to the basic search, but it allows restriction to specific data types. The second type of advanced query is more specific of Selectome. The user must select a branch from the taxonomy, and specify whether to search for branches with or without positive selection, and whether these are speciation branches or duplication branches. For example a query for positive selection on the mammalian branch (represented by Theria in Selectome release 1) without gene duplication returns 74 gene families.
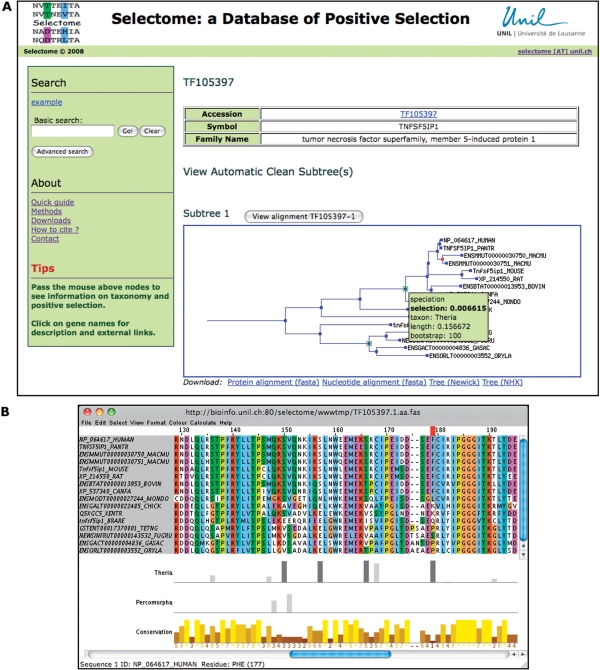
The main view presents a gene family. It contains annotations from TreeFam, and information on positive selection (Figure 2). For each subtree, the phylogeny is represented using a customized version of the TreeFam API. In addition to default color codes of TreeFam, a green box marks branches where positive selection was detected. Passing the mouse above such a node highlights the corresponding P-value for the likelihood test. Above each subtree is a link to the protein alignment, which launches the Jalview Applet (16). The Applet automatically uploads an annotation track per branch with positive selection, which contains the PP for sites detected under positive selection. These PP values are represented as bars of height proportional to their value. In addition, only sites with a PP ⩾ 99% are represented by a black bar, while others are represented by shades of gray (light gray if 50% ⩽ PP < 95%; dark gray if 95% ⩽ PP < 99%). The user may take advantage of the tools of Jalview, e.g. to color only sites with strong support for positive selection on a branch of interest.
Figure 2.
Screenshot of the view of positive selection on family TF105397 (tumor necrosis factor superfamily, member 5-induced protein 1). (A) Default gene family view. Green boxes on tree nodes indicate positive selection on the corresponding branch. Information on the mammalian branch is shown by passing the mouse above the corresponding node. (B) Protein alignment view. Corresponding annotation tracks under the protein alignment indicate positively selected sites. Site 179 of the alignment has been selected; it shows the fixation of phenylalanine in most mammals. Screenshots from Selectome release 1.
CONCLUSIONS AND PERSPECTIVES
Selectome provides access to results from the most rigorous test for positive selection, the branch-site likelihood test as improved by Zhang et al. (8). Using this test in combination with q-value correction for multiple testing has been shown to provide accurate results with a low rate of false positives (2,7). Selectome provides an intuitive Web interface, which allows easy access to all results. Important features are the mapping of positive selection on branches of the phylogenetic tree, and on sites of the multiple alignment. To our knowledge, no other resource provides such intuitive access to the results of advanced tests of positive selection. This is all the more relevant that the original software package, PAML (6), while very complete and powerful, is also difficult to use for many biologists. We must note here our debt to the great development work done in the TreeFam (10) and Jalview (16) projects. We expect Selectome to be useful both for functional and evolutionary studies. Notably, genetic or genomic studies may be enhanced by identifying sites which have potentially been the target of functional shifts in specific lineages. This could either lead to a note of caution concerning the conservation of function between orthologs, or to the identification of sites of interest for further investigation.
One other resource provides positive selection information mapped on the tree, TAED (17). TAED does not use a stringent likelihood test, nor a correction for multiple testing, and does not provide information on sites under positive selection, but only a global dN/dS per branch. The only other database to make use of the branch-site test, to our knowledge, is the Human PAML Browser (18). But that resource presents results without any correction for multiple testing, and without a graphical Web interface. Also, Selectome covers a diversity of phylogenetic branches, whereas the Human PAML Browser is limited to the branch leading to humans. Selecton (19) is a webservice for positive selection, which provides several sophisticated models. Selecton is not a database, and does not allow querying precomputed results. Thus we feel that Selectome represents a significant contribution.
The future development of Selectome will follow two main directions. First, as data coverage increases, it will be possible to apply our methodology to more lineages. We plan to update Selectome with each new release of TreeFam. While Selectome release 1 is based only on the more reliable TreeFam-A gene families, future releases will also include TreeFam-B in the interest of completeness. Second, research on methods to detect positive selection is ongoing (3,4). We plan to implement other methods into Selectome, but only once their use and limitations have been thoroughly established. Indeed Selectome does not aim to be a repository of all possible selection computations, but a high quality resource for trustworthy information on positive selection.
FUNDING
Etat de Vaud; Swiss National Science Foundation (116798). Funding for open access publication charge: Etat de Vaud.
Conflict of interest: None declared.
ACKNOWLEDGEMENTS
The computations were performed at the Vital-IT (http://www.vital-it.ch) centre for high-performance computing of the Swiss Institute of Bioinformatics. We thank James Procter of the Jalview development team for his help.
REFERENCES
- 1.Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol. Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 2.Studer RA, Duret L, Penel S, Robinson-Rechavi M. Pervasive positive selection on duplicated and non duplicated vertebrate protein coding genes. Genome Res. 2008;18:1393–1402. doi: 10.1101/gr.076992.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen JD, Wong A, Aquadro CF. Approaches for identifying targets of positive selection. Trends Genet. 2007;23:568–577. doi: 10.1016/j.tig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Anisimova M, Liberles DA. The quest for natural selection in the age of comparative genomics. Heredity. 2007;99:567–579. doi: 10.1038/sj.hdy.6801052. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z. Computational Molecular Evolution. USA: Oxford University Press; 2006. [Google Scholar]
- 6.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 7.Anisimova M, Yang Z. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol. Biol. Evol. 2007;24:1219–1228. doi: 10.1093/molbev/msm042. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Coghlan A, Ruan J, Coin LJ, Heriche J.-K, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan J, Li H, Chen Z, Coghlan A, Coin LJM, Guo Y, Heriche J.-K, Hu Y, Kristiansen K, Li R, et al. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J. Frequent False Detection of Positive Selection by the Likelihood Method with Branch-Site Models. Mol. Biol. Evol. 2004;21:1332–1339. doi: 10.1093/molbev/msh117. [DOI] [PubMed] [Google Scholar]
- 14.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 16.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 17.Roth C, Betts MJ, Steffansson P, Saelensminde G, Liberles DA. The Adaptive Evolution Database (TAED): a phylogeny based tool for comparative genomics. Nucleic Acids Res. 2005;33:D495–D497. doi: 10.1093/nar/gki090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickel GC, Tefft D, Adams MD. Human PAML browser: a database of positive selection on human genes using phylogenetic methods. Nucleic Acids Res. 2008;36:D800–D808. doi: 10.1093/nar/gkm764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007;35:W506–W511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]