Abstract
ATTED-II (http://atted.jp) is a database of gene coexpression in Arabidopsis that can be used to design a wide variety of experiments, including the prioritization of genes for functional identification or for studies of regulatory relationships. Here, we report updates of ATTED-II that focus especially on functionalities for constructing gene networks with regard to the following points: (i) introducing a new measure of gene coexpression to retrieve functionally related genes more accurately, (ii) implementing clickable maps for all gene networks for step-by-step navigation, (iii) applying Google Maps API to create a single map for a large network, (iv) including information about protein–protein interactions, (v) identifying conserved patterns of coexpression and (vi) showing and connecting KEGG pathway information to identify functional modules. With these enhanced functions for gene network representation, ATTED-II can help researchers to clarify the functional and regulatory networks of genes in Arabidopsis.
INTRODUCTION
Genes involved in related biological pathways are often expressed cooperatively, and thus information on their coexpression is key to understanding biological systems (1). Indeed, coexpression data have been applied to a wide variety of experimental approaches, such as gene targeting, regulatory investigations and/or identification of potential partners in protein–protein interactions (PPIs) (2,3). Large amounts of gene expression data are required to reliably predict coexpressed gene relationships. Toward this end, DNA microarrays have provided vast amounts of gene expression data that are stored in several public repositories (4–7). Using these large stores of public data, several coexpression databases have been constructed and are widely used, especially in Arabidopsis research (8–14). Although these databases provide similar information, each has unique functionalities that enhance the potential discovery of gene coexpression, such as cis element findings cooperated with coexpressed gene selection (10), combinatorial analyses using gene coexpression from multiple species (14), or an interactive user interface using Java technology (9).
One such coexpression database for Arabidopsis, ATTED-II, has the unique aspect of network representation of gene coexpression in addition to a simple gene list representation (12). Network representation can provide relationships between functional modules along with each gene-to-gene relationship, and thus it can yield a systematic understanding of the target species. ATTED-II previously provided three types of coexpressed gene networks, namely the network of coexpressed genes for every locus, that of the genes with the same gene ontology (GO) annotation (15), and that of any user-defined set of genes. However, the coexpressed gene networks in the previous ATTED-II had some limitations, denoted A and B as follows. (A) When large numbers of nodes and edges were involved, the networks became huge and the graphical representation was messy. (B) The understanding of gene networks was highly dependent on the user's knowledge of target phenomena, due to the lack of intuitive interfaces for appropriate annotations.
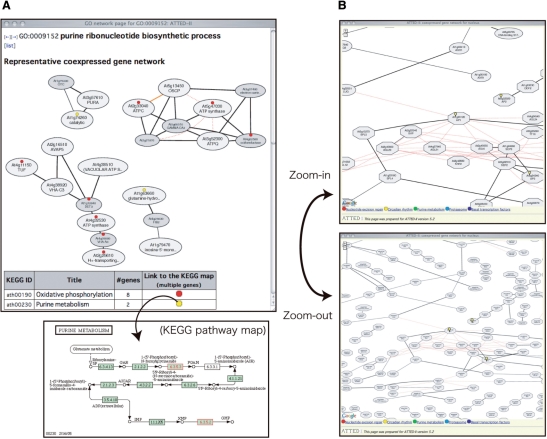
In this article, we describe new features of ATTED-II implemented after the previous publication (12) to address points A and B as follows. (A-1) A new coexpression measure was developed to retrieve functionally related genes more accurately. (A-2) Clickable maps were implemented for all gene networks to allow step-by-step navigation of the gene map. (A-3) Google Maps API was introduced to represent large networks interactively. (B-1) Known PPIs were added to confirm the regulation of protein levels. (B-2) Conserved coexpression information was added to identify the core components of functional modules. (B-3) KEGG annotation information (16) was added to identify functional networks. (B-4) Transcription factor gene annotation was added. Details of the new features are described in the following sections, along with examples of the new coexpressed gene networks for genes with a specific GO annotation (Figure 1A) and for genes whose products function in a specific organelle (Figure 1B). The history of ATTED-II, as well as other miscellaneous updates, is shown in Table 1.
Figure 1.
(A) An example of coexpressed gene networks for ‘purine ribonucleotide biosynthetic process’ (GO:0009152). Solid edges (lines) indicate gene coexpression, and red dotted edges indicate PPIs. Orange edges indicate conserved coexpression between Arabidopsis and at least one of three mammalian species (human, mouse and rat). Octagon-shaped nodes indicate TF genes, and circular nodes indicate other types of genes. Large nodes are genes with the GO annotation, which are the input genes used to construct these networks. The small gray-shaded nodes are genes automatically selected based on the strength of gene coexpression. Common KEGG pathways in the network are denoted by color-coded (red or yellow) dots in the nodes. (Note that this KEGG pathway is just part of the whole pathway.) This page can be obtained at [http://atted.jp/data/gonetwork/GO:0009152.html]. (B) An example of coexpressed gene networks for an organelle (nucleus). We used Google Maps API to display the huge coexpression map interactively. Yellow balloons mark the user-specified genes (AP1, AP2, AP3 in this case). Black solid edges indicate gene coexpression, and red dotted edges indicate PPIs. Octagon-shaped nodes indicate TF genes, and circular nodes indicate other types of genes. This page can be generated at [http://atted.jp/gmap/nucleus/?gene=At1g69120,At5g52020,At3g54340]. The window focuses on the first gene (At1g69120).
Table 1.
Evolution of ATTED-II versions over the last 2 years
| version | Date | Gene coexpression | Gene model | Other annotations |
| ver. 5.2 | 2008.07.19 | ↑ | TAIR8 | updated |
| ver. 5.1 | 2008.04.08 | ver. c4.1 | ↑ | ↑ |
| ver. 5.0 | 2008.03.18 | ver. c4.0 | ↑ | updated |
| ver. 4.4 | 2007.10.16 | ↑ | ↑ | updated |
| ver. 4.3 | 2007.09.12 | ver. c3.1 | TAIR7 | updated |
| ver. 4.2 | 2006.07.24 | ↑ | ↑ | updated |
| ver. 4.1 | 2006.06.03 | ↑ | ↑ | updated |
| ver. 4.0 | 2006.05.25 | ver. c3.0 | TAIR6 | updated |
↑: same condition as previous version. Details of the updates of ‘Other annotations’ are available from http://atted.jp/versions.shtml
IMPROVEMENTS IN GENE NETWORK CONSTRUCTION AND REPRESENTATION
A new coexpression measure
Pearson's correlation coefficient (PCC) is widely used to assess the extent of gene coexpression, and we previously used PCC weighted by sample redundancies (12). However, we found that many functionally relevant coexpressed gene pairs have low PCCs. To avoid discarding potentially important coexpressed gene pairs having low PCCs, we introduced a new measure of gene coexpression, Mutual Rank (MR), that is calculated as the geometric mean of the correlation rank of gene A to gene B and of gene B to gene A. Lists of coexpressed genes based on MR values are now provided, in which a weighted PCC for each gene pair is also presented to compare PCCs with coexpression data provided in previous versions of ATTED-II or in other databases. All versions of gene coexpression data are available at http://atted.jp/top_search.shtml#coexversion.
Coexpressed gene network in ATTED-II
To generate coexpressed gene networks, the three genes having the highest MR are connected with each other. For simplicity, the gene network presented on the locus page of ATTED-II is limited to 21 nodes. Three types of edges (lines) are used to draw the networks, that is, bold edges (MR <5), normal edges (5 ≤ MR < 30) and thin edges (MR ≥30) (Figure 1). Each node is clickable to visit the corresponding locus in the network. Table 2 shows the current statistics of the coexpressed gene networks in ATTED-II.
Table 2.
Statistics of gene networks provided in ATTED-II
| Number of pages for coexpressed gene network |
Network size |
||||||
|---|---|---|---|---|---|---|---|
| Type of page | Total | With PPI annotation | With conserved coexpression | With common KEGG pathway | Min | Mean | Max |
| Locus | 20 876 | 517 | 758 | 4399 | 9 | 21 | 21 |
| GO term | 818 | 142 | 71 | 297 | 3 | 19 | 196 |
| Organelle | 6 | 6 | 5 | 6 | 25 | 467 | 1161 |
Coexpressed gene networks for organelles interfaced with Google Maps
Cellular organelles have distinct functions, and thus each organelle can serve as a unique research/pharmacological target. In particular, the chloroplast is an important target for many plant researchers. We constructed a large network of coexpressed genes for each Arabidopsis organelle to focus on their functional relationships. Genes encoding proteins that function in each organelle were identified by the following GO annotations: chloroplast (GO:009507), nucleus (GO:005634), mitochondrion (GO:005739), golgi (GO:005794), endoplasmic reticulum (GO:005783) and vacuole (GO:005773). All of the children terms until the end of GO hierarchy are also included for the six organelle. The global map for an organellar coexpressed gene network is too large to be visualized as a single image on a static page, and thus we used Google Maps API (Application Programming Interface, http://www.google.com/apis/maps/) to navigate large coexpression networks interactively (Figure 1B). This interface allows visualization of all gene relationships for each organelle (http://atted.jp/browsing/list_organelle.html).
IMPROVEMENTS IN GENE NETWORK ANNOTATIONS
Conserved coexpression as an edge annotation
Coexpression of certain sets of Arabidopsis genes is conserved across a broad range of species. Such ‘conserved coexpression’, as we call it here, can predict gene-to-gene functional relationships more accurately because their coexpression has been conserved across evolution (17). Therefore, the new ATTED-II defines/shows conserved coexpression by a colored edge in coexpressed networks (Table 2; orange edges in Figure 1). Conserved coexpression was assigned if a gene pair coexpressed in Arabidopsis (three genes having the highest MR for each gene) was also coexpressed in one of three organisms, namely human, mouse or rat, for which coexpression data were obtained from COXPRESdb (13) and ortholog information from HomoloGene (18). At present, the conserved coexpressions do not cover plant-specific functions because microarray data, which are plentiful for mammals, are largely unavailable for plant species other than Arabidopsis. When microarray analyses for non-Arabidopsis plants begin to accumulate in public databases, we will add coexpression data for other plants, which will afford the opportunity to identify core components of functional modules among plant species.
Integration of PPI networks and coexpressed gene networks
Although gene coexpression and PPIs are well correlated in prokaryotes, the correlation is not as strong in eukaryotes (19), probably because the layer of regulatory mechanisms is different between coexpression and PPI. In other words, gene coexpression reflects transcriptional/RNA-level regulation, whereas PPIs reflect protein-level regulation. Thus, PPI information can serve to complement, and thereby enhance, the reliability of predicted gene-to-gene functional relationships. Therefore, in the new ATTED-II we added PPI information from TAIR (7) and IntAct (20) databases (Table 2). In Figure 1A, the PPIs (red dotted edges) basically agree with the coexpression to connect genes in the same gene cluster. On the other hand, in Figure 1B, one of the PPIs connects distinctive coexpressed gene groups, possibly indicating that PPIs complement gene coexpression to more accurately represent gene-to-gene functional structures. The annotations of conserved coexpression and PPI along with the coexpression strength (MR and PCC) for query gene pairs are available on the ATTED-II search page (http://atted.jp/top_search.shtml#edgeannotation). Despite the usefulness of PPI data, experimentally verified PPI data in Arabidopsis are limited. Thus, in future versions of ATTED-II we plan to include prediction-based PPIs using reported methods (21,22).
KEGG pathways for gene annotation
To help interpret coexpression module functions, the nodes are marked with KEGG pathway annotations (16). For each gene network, up to five KEGG pathways with relatively large numbers of genes are selected (Table 2; Figure 1A). For example, when genes with the GO annotation ‘purine ribonucleotide biosynthetic process’ (GO:0009152) are used as input queries for network construction, they are represented as large nodes, and genes that are strongly coexpressed with the input genes are automatically picked and added to the network and are represented as small gray-shaded nodes (Figure 1A). As a result, four coexpressed gene networks emerged in this example. Based on the KEGG annotation, two of them are related to ‘oxidative phosphorylation’ (ath00190), and the others are involved in ‘purine metabolism’ (ath00230). It should be noted that some genes that are selected by coexpression (smaller darkly shaded nodes in Figure 1A) are also marked by the same KEGG pathways. This suggests that the gene coexpression data in ATTED-II can correctly select functionally related genes even though they lack GO annotations.
The same marks in the table, placed just below the network, represent links to the KEGG pathway, in which the marked genes in the coexpressed gene networks are highlighted by red boxes in the KEGG metabolic pathway. This allows coexpressed gene modules to be associated with metabolic pathways.
Annotation of transcription factors
To investigate regulatory relationships, transcription factor genes are represented as octagon-shape nodes (Figure 1A). Coexpression of a gene(s) with a transcription factor gene(s) can provide clues about regulatory mechanisms (23). Annotations of transcription factors were obtained from AGRIS (24). We are also investigating annotations from other transcription factor databases (25,26), which may be incorporated to ATTED-II in the future.
OTHER IMPROVEMENTS
Coexpression viewer
In ATTED-II, data from many experiments are simultaneously used to calculate PCC, and thus the meaning of the contribution of each microarray sample is not clearly discernable in the final coexpression values. Therefore, we prepared a coexpression viewer to clarify the contribution of each microarray sample to the deduced coexpression for any gene pair. The coexpression viewer provides two views: scatter plots constructed from data representing the two gene expression patterns, and the contribution of each experimental aspect to the expression pattern similarity. In both views, the samples are divided into ‘developmental samples’ and ‘others’ because the former causes more dynamic expression changes and thus may mask the effects of the latter.
To focus on the effects of the non-developmental samples more directly, coexpression values without developmental samples are also available at http://atted.jp/top_search.shtml#coexversion, as coexpression data version c6.0.
Version numbering
ATTED-II is composed of our own coexpression data and integrated public annotations. Coexpression data are updated yearly, and public annotations are updated every few months (Table 1). Each data update is assigned a new version number to prevent confusion about the data update and as a reference for users’ publications. The major version number essentially corresponds to the coexpression data update, whereas the minor version number corresponds to a public data update. Version histories can be checked at http://atted.jp/versions.shtml. Note that we maintain previous versions of gene coexpression data, but we do not store the any other annotations.
FUNDING
This work was partially supported by a Grant-in Aid for Scientific Research on the Priority Area ‘Transportsome’ and by the Global COE Program (Center of Education and Research for Advanced Genome-based Medicine) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charges: Annual budget for Human Genome Center, Institute of Medical Science, The University of Tokyo.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, The University of Tokyo.
REFERENCES
- 1.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48:381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- 3.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput. Biol. 2007;3:e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M, et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinhauser D, Usadel B, Luedemann A, Thimm O, Kopka J. CSB.DB: a comprehensive systems-biology database. Bioinformatics. 2004;20:3647–3651. doi: 10.1093/bioinformatics/bth398. [DOI] [PubMed] [Google Scholar]
- 9.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008;2008:420–747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 11.Manfield IW, Jen CH, Pinney JW, Michalopoulos I, Bradford JR, Gilmartin PM, Westhead DR. Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 2006;34:W504–W509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Res. 2008;36:D77–D82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mutwil M, Obro J, Willats WG, Persson S. GeneCAT–novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 2008;36:W320–W326. doi: 10.1093/nar/gkn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhardwaj N, Lu H. Correlation between gene expression profiles and protein-protein interactions within and across genomes. Bioinformatics. 2005;21:2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- 20.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145:317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui J, Li P, Li G, Xu F, Zhao C, Li Y, Yang Z, Wang G, Yu Q, Li Y, et al. AtPID: Arabidopsis thaliana protein interactome database–an integrative platform for plant systems biology. Nucleic Acids Res. 2008;36:D999–D1008. doi: 10.1093/nar/gkm844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 26.Riano-Pachon DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: an integrative plant transcription factor database. BMC Bioinform. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]