Abstract
Molecular interaction data exists in a number of repositories, each with its own data format, molecule identifier and information coverage. Michigan molecular interactions (MiMI) assists scientists searching through this profusion of molecular interaction data. The original release of MiMI gathered data from well-known protein interaction databases, and deep merged this information while keeping track of provenance. Based on the feedback received from users, MiMI has been completely redesigned. This article describes the resulting MiMI Release 2 (MiMIr2). New functionality includes extension from proteins to genes and to pathways; identification of highlighted sentences in source publications; seamless two-way linkage with Cytoscape; query facilities based on MeSH/GO terms and other concepts; approximate graph matching to find relevant pathways; support for querying in bulk; and a user focus-group driven interface design. MiMI is part of the NIH's; National Center for Integrative Biomedical Informatics (NCIBI) and is publicly available at: http://mimi.ncibi.org.
INTRODUCTION
Both the volume and number of data sources in molecular biology are increasing rapidly. Often multiple resources provide overlapping, partial and polymorphic views of the same data. Scientists often wish to piece together data from multiple source databases. Many web sites today recognize this need and provide one-stop access to multiple external databases. These include (1–4) to name a few. The IMEx consortium (http://imex.sf.net/) facilitates sharing of interaction information across multiple databases maintained by independent curators. However, all of these databases perform ‘shallow integration’: they make access convenient for the user by making all data available at one place, but make no attempt to pull that data into a cohesive whole. Source databases have overlapping coverage, resulting in multiple entries of the same information in the integrated result. For example, the same source publication may be cited in the interaction record in DIP, BIND, HPRD and BioGRID.
Michigan molecular interactions (MiMI) helps scientists search through large quantities of information by integrating all information from participating data sources through the process of deep merging. As a result, redundant data are removed and related data are combined. Furthermore, in doing so, MiMI keeps track of the ‘provenance’ of each piece of information, or from where it was obtained. A website with this functionality was launched about 2 years ago, and described in (5). Since then, we have received a great deal of feedback, and have made further progress in integrating information. A completely redone MiMI Release 2 (MiMIr2) is now being released, and is the subject of this article. Noteworthy new features are mentioned below.
MiMI is a component of the NIH's; National Center for Integrative Biomedical Informatics (http://www.ncibi.org), and is available at: http://mimi.ncibi.org free of charge, and with no registration required. A dump of MIMIr2 data is also available for free, but comes with some limitations on use and requires a license agreement.
BIOLOGICAL CONCEPTS
A central need for many scientists is to place a protein interaction in context—either in terms of genes that code for these proteins or in terms of pathways of which this interaction may be a part. MiMIr2 facilitates this by integrating relevant data from NCBI Entrez Gene on the one hand, and Reactome and KEGG on the other; and also by providing novel graph browsing and graph searching capabilities (described in the next section).
MiMIr1 had information primarily regarding protein interactions, designed for scientists to query based on proteins of interest. We found, however, that most users had genes rather than proteins of interest. We also found many users cursory in mapping from genes of interest to proteins of interest—it is tempting to take the known gene name, assume that it codes for exactly one protein, and that the protein it codes for has the same name as the gene. We all know this is not always the case, but we do it nonetheless, getting results that are less than satisfactory. MiMIr2 remains an interaction database, but uses genes rather than proteins as the central identifying entity. Users ask about relationships between genes, and are informed about interacting products of these genes.
One important purpose of looking at protein (or gene) interactions is to determine biological pathways of interest in which the subject genes plays a role. MiMIr2 has imported pathways from KEGG (6) and Reactome (7). For each gene and for each interaction, MiMIr2 can be used to find pathways in which it participates.
IDENTITY
The issue of identity is determining when two database entries refer to the same real world object. If two proteins have an almost identical sequence of amino acids and are expressed in the same organism, then they are likely to be the same. MiMIr1 included many rules, such as this, to figure out when two databases referred to the same protein. While this worked well for the most part, we found many protein fragments, and other variants, were recorded as separate molecules. When a scientist queried for a particular protein, these variants would not be returned, since they were considered distinct entities.
Given the introduction of gene as the primary query entity in MiMIr2, we had a natural way to address this vexing identity question by mapping proteins to their coding genes. Now, two variants, even if treated as distinct proteins, can still be associated with the same gene. When a user queries for interactions associated with this gene, all proteins and fragments, and their interactions, are returned.
QUERY SPECIFICATION
MiMIr1 provided a wide variety of query interfaces, including a form-based interface and a visual query builder. However, we found that most users had a strong preference for an unfielded ‘Google-style’ search box into which they could enter query terms of their choice and then sift through returned results. (users also liked point and click query specifications—see next section on Cytoscape). As such, MiMIr2 has simplified its query interface to provide users with a single query box. The only field requiring explicit specification is the organism of interest, if there is one. Users can enter anything at all, including words used in a textual description field, including associations with a disease or biological process or molecular function. To support such queries better, we associate GO (8) labels with MiMIr2 entries.
This resulted in a very useful query facility. However, there frequently are a number of genes associated with a biological term. Our system would order these based on information retrieval metrics, which may not match a scientist's; view of relative importance. To help address this, we developed a tool, Gene2Mesh, which could be used to go from a MeSH (9) heading to a ranked list of genes mentioned frequently in papers to which that MeSH heading was associated. It could also go from a Gene to a ranked list of MeSH headings associated with the gene. We integrated Gene2Mesh with MiMI so that a user can refine their topical query with MeSH terms if an initial attempt through an open text box yields unsatisfactory results.
Gene2Mesh uses occurrence frequency to estimate importance of connection. However, not all occurrences are equally important. If we wish to find genes that are central to a particular disease or process, we should expect them to be centrally located in a graph of genes related to that disease or process. This notion of graph centrality is exploited to rank genes in a new tool, GIN (10,11), part of CLAIRLIB (12). MiMIr2 also integrates GIN, so that users can choose genes of importance related to a specific biological concept. Note that Gene2Mesh and GIN provide a means for users to learn about new genes related to a concept of interest.
It is natural, when specifying queries, to consider one gene at a time. Clicking through from query results also works one click, to a specific gene, at a time. Yet, there is a class of scientists who are interested in sets of genes, for example, a set that is differentially over-expressed in some microarray experiment. In MiMIr2, we added a set-of-genes functionality to support such users. Users can either type in a list of gene symbols or gene IDs, or import a file containing these, and use that to query MiMIr2 through a special interface set up explicitly for this purpose. Once they are past this step, everything else works just as for single gene specifiers.
VIEWING INFORMATION
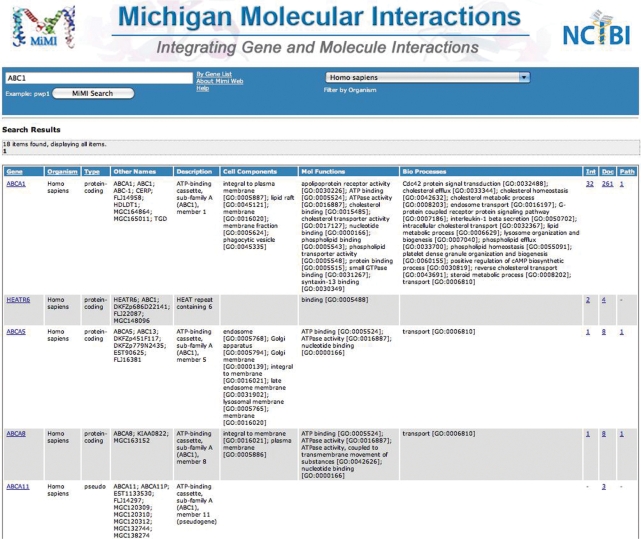
We observed scientists using MiMIr1, and also conducted focus groups, to identify result presentation features that users liked and disliked. Based on this feedback, we created a very sparse, but maximally informative, user interface, with every screen laid out carefully, and designed to work whether the number of returned results is 2 or 2000. By making the front page simple and minimalist in MiMIr2, we invite the user to type anything and get started with viewing useful results. Figure 1 shows the results of a query for the gene ABC1. The user is presented upfront with information, such as organism, aliases, description, GO cellular component, GO molecular function and GO biological process in order to facilitate identification and confirmation by the user. If the user determines this to be the gene of interest, quick links to further gene and protein information, interaction information, pathways and documentation exist for quick and easy navigation.
Figure 1.
The new results page in MiMI r2.
Sometimes a user may just wish to browse the database. Once a gene of interest is located, for example, a scientist may want to look around and see what else is there ‘in the neighborhood’. To support such use, we developed a browsing interface. The user can look through sets of genes that have the same values for one or more of several attributes.
Additionally, many scientists find a visual interface useful to see a graph of interactions. Cytoscape (13) is a popular tool widely used for this purpose. MiMIr1 permitted users to export lists of interactions in SIF format, which could be read and viewed in a Cytoscape browser. However, there were several limitations: output storage, cytoscape initialization, format limitations, inability to query-on-the-fly with MiMI, etc. All of these issues have been addressed in MiMIr2 through the creation of a MiMI plugin for Cytoscape (14) and through the development of a single-click web start version of Cytoscape that can be invoked from the MiMI web site. Most pages in the MiMI web site now have a clickable link to Cytoscape that will lead the user to a Cytoscape browser session. Users starting from Cytoscape now have access to MiMI data, including all gene and interaction attributes, so that they can query based on these while remaining in the Cytoscape environment. The MiMI plugin for Cytoscape has already been downloaded 2318 times since its release earlier this year.
Cytoscape is a large software package with many features. While we know many scientists who absolutely love it, we found others who have never used it and found the learning curve too steep to invest the effort required to become effective users. To address this section of our user base, we developed a stripped down ‘NetBrowser’ in Adobe Flash, with extremely limited functionality, but with a very simple interface that any one can use without training. From most MiMIr2 pages, the user can get to NetBrowser and see a set of interactions visually rather than in tabular form.
Pathway databases have traditionally had their own visual representations of pathways, and we have preserved these as we integrated pathway data in MiMIr2. Moreover, we have provided a unique pathway search capability through SAGA (15,16). Once the user has identified a set of interacting genes of interest in NetBrowser, this interacting set can be used to query for pathways in which similar interaction patterns of these genes can be found (by clicking the button, ‘Export to SAGA’ from within NetBrowser).
DATA VERIFICATION
Scientists typically want to understand where data came from before they are willing to trust it. Being cognizant of this, in MiMIr1, we carefully preserved provenance information regarding the source of every piece of data, and we continue to do this in MiMIr2. Many interaction databases provide them with PubMed IDs of publications from which a fact was derived (and this information was preserved in MiMIr1). The user is then free to obtain the original publication and peruse it to determine the reliability of the reported interaction.
MiMIr2 provides a unique capability, grounded in natural language processing, for the user to look up, while in MiMIr2 itself, the specific sentences in cited publications from which the interaction information was derived, with extracted terms of importance highlighted. (After search, click on the Gene name to see gene detail; scroll to Related Documents and click on View.) Figure 2 shows a sample of the available information. The user is thus saved the effort of locating and reading the entirety of the cited publication—rather their attention is immediately drawn to the few sentences that are likely to matter the most.
Figure 2.
Natural language processing identification of sentences within a document dealing with information of interest.
DATA SETS
MiMI currently has over 3.7 million interactions, along with information about approximately 3.5 million genes, 19.2 million molecules and 1288 pathways. Table 1 contains the list of sources in MiMI and their contributions. Additionally, supplementary protein information was integrated from: GO (8), InterPro (26), IPI (27), miBLAST (28), OrganelleDB (29), OrthoMCL (30) PFam (31) and ProtoNet (32).
Table 1.
Interaction and pathway datasets in MiMIr2
| Source | Num. Mol. | Num. Int. |
|---|---|---|
| BIND (17) | 111 112 | 233 201 |
| Center for Cancer Systems Biology, Harvard (18) | 3134 | 6683 |
| BioGRID (19) | 48 855 | 167 330 |
| DIP (20) | 76 771 | 49 677 |
| HPRD (21) | 73 209 | 116 333 |
| IntAct (22) | 197 141 | 77 780 |
| Max Delbrück Center (23) | 1909 | 3269 |
| MINT (24) | 195 119 | 125 672 |
| Reactome (7) | 48 808 | 2 375 780 |
| WSU Campylobacter Jejuni Interactome (25) | 1332 | 12 012 |
FUNDING
National Institutes of Health (R01 LM008106 and U54 DA021519, partial). Funding for open access charge: NIH grant U54 DA021519.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liebel U, Kindler B, Pepperkok R. Bioinformatic ‘Harvester’: a search engine for genome-wide human, mouse, and rat protein resources. Methods Enzymol. 2005;404:19–26. doi: 10.1016/S0076-6879(05)04003-6. [DOI] [PubMed] [Google Scholar]
- 3.Birkland A, Yona G. The BIOZON Database: a hub of heterogeneous Biological Data. Nucleic Acids Res. 2006;34:D235–D242. doi: 10.1093/nar/gkj153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson SB, Crabtree J, Brunk BP, Schug J, Tannen V, Overton GC, Stoeckert CJ., Jr K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Syst. J. 2001;40:512–531. [Google Scholar]
- 5.Jayapandian M, Chapman A, Tarcea VG, Yu C, Elkiss A, Ianni A, Liu B, Nandi A, Santos C, Andrews P, et al. Michigan Molecular Interactions (MiMI): Putting the Jigsaw Puzzle Together. Nucleic Acids Res. 2007;35:D566–D571. doi: 10.1093/nar/gkl859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi-Tope G, Gillespie M, Vastrik I, D'E;ustachio P, Schmidt E, deBono B, Jassal B, Gopinath G, Wu G, Matthews L, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gene Ontology Consortium. Creating the Gene Ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sewell W. Medical subject headings in MEDLARS. Bull. Med. Lib. Assoc. 1964;52:164–170. [PMC free article] [PubMed] [Google Scholar]
- 10.Özgür A, Vu T, Erkan G, Radev DR. Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics. 2008;24:277–285. doi: 10.1093/bioinformatics/btn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leitner F, Krallinger M, Rodriguez-Penagos C, Hakenberg J, Plake C, Kuo C-J, Hsu C-N, Tasi RT-H, Hung H-C, Lau WW, et al. Introducing meta-services for biomedical information extraction. Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-s2-s6. (Suppl. 2), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radev DR, Hodges M, Fader A, Joseph M, Gerrish J, Schaller M, dePeri J, Gibson B. Technical Report CSE-TR-536-07. University of Michigan, Department of Electrical Engineering and Computer Science; 2007. CLAIRLIB Documentation v1.03. [Google Scholar]
- 13.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Ade AS, Tarcea VG, Weymouth TE, Mirel BR, Jagadish HV, States DJ. Integrating and annotating the interactome using the MiMI plugin for Cytoscape. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn501. [Epub ahead of print; doi: 10.1093/bioinformatics/btn501; 23 September] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian Y, McEachin RC, Santos C, States DJ, Patel JM. SAGA: a subgraph matching tool for biological graphs. Bioinformatics. 2007;23:232–239. doi: 10.1093/bioinformatics/btl571. [DOI] [PubMed] [Google Scholar]
- 16.Tian Y, Patel JM. International Conference on Data Engineering. Cancún, México: 2008. [Google Scholar]
- 17.Bader G, Betel D, Hogue CWV. BIND: the biomolecule interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han J-DJ, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJM, Cusick ME, Roth FP, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 19.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xenarios I, Salwínski Ł, Duan XJ, Higney P, Kin S.-M, Eisenberg D. DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TKB, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B, Roepstorff P, Valencia A, et al. IntAct - an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stelzl U, Worm U, Lalowski M, Hänig C, Brembeck FH, Göhler H, Strödicke M, Zenkner M, Schönherr A, Köppen S, et al. a human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 24.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrish J, Yu J, Liu G, Hines J, Chan J, Mangiola B, Zhang H, Pacifico S, Fotouhi F, DiRita V, et al. A Proteome-wide Protein Interaction Map for Campylobacter Jejuni. Genome Biology. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The international protein index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 28.Kim YJ, Boyd A, Athey BD, Patel JM. miBLAST: scalable evaluation of a batch of nucleotide sequence queries with BLAST. Nucleic Acids Res. 2005;33:4335–4344. doi: 10.1093/nar/gki739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiwatwattana N, Kumar A. Organelle DB: a cross-species database of protein localization and function. Nucleic Acids Res. 2005;33:D598–D604. doi: 10.1093/nar/gki071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F, Mackey AJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, Mistry J, Schuster-Báockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. PFam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasson O, Vaaknin A, Fleischer H, Portugaly E, Bilu Y, Linial N, Linial M. ProtoNet: hierarchical classification of the protein space. Nucleic Acids Res. 2003;31:348–352. doi: 10.1093/nar/gkg096. [DOI] [PMC free article] [PubMed] [Google Scholar]