Abstract
The Mouse Genome Database (MGD, http://www.informatics.jax.org/), integrates genetic, genomic and phenotypic information about the laboratory mouse, a primary animal model for studying human biology and disease. Information in MGD is obtained from diverse sources, including the scientific literature and external databases, such as EntrezGene, UniProt and GenBank. In addition to its extensive collection of phenotypic allele information for mouse genes that is curated from the published biomedical literature and researcher submission, MGI includes a comprehensive representation of mouse genes including sequence, functional (GO) and comparative information. MGD provides a data mining platform that enables the development of translational research hypotheses based on comparative genotype, phenotype and functional analyses. MGI can be accessed by a variety of methods including web-based search forms, a genome sequence browser and downloadable database reports. Programmatic access is available using web services. Recent improvements in MGD described here include the unified mouse gene catalog for NCBI Build 37 of the reference genome assembly, and improved representation of mouse mutants and phenotypes.
INTRODUCTION
The Mouse Genome Database (MGD) is a comprehensive public resource providing integrated access to genetics, genomics, functional and phenotypic data for the laboratory mouse (1–3). MGD is a core database component of the Mouse Genome Informatics (MGI) database resource (http://www.informatics.jax.org). Other resources that are integrated with MGD as part of the MGI resource include the Gene Expression Database (GXD) (4), the Mouse Tumor Biology Database (MTB) (5) and the Gene Ontology (GO) project (6).
MGD facilitates translational biomedical research via a comprehensive database resource integrated with bio-ontological semantic standards that enhances the use of the laboratory mouse as a model animal system for studying human biology. Primary data types in MGD include sequences, genetic and physical maps, genes, gene function, gene families, strains, mutant phenotypes, SNPs, animal models of human disease and mammalian homology. MGD annotations are integrated through a combination of expert human curation and automated processes. Examples of vocabularies and ontologies utilized in MGD include the GO (6), Mammalian Phenotype (MP) Ontology (7) and the Anatomical Dictionary of Mouse Development (8). Mouse genes and gene products in MGD are also associated with multiple other informatics resources including the Online Mendelian Inheritance in Man (OMIM), UniProt protein resources and PIR protein super family classifications. MGI is the authoritative source for mouse gene and strain nomenclature and GO functional annotations. MGI is the most comprehensive public resource of information on mouse phenotypes and associations between mouse models and human disease.
Data in MGD are updated daily. Data access is accomplished via dynamically generated web pages, text files available via FTP (updated nightly) and through direct SQL (account is required). In general, there are 4–6 major software releases per year to support access and display of new data types. A recent summary of MGD content is shown in Table 1.
Table 1.
Snapshot of data content in MGD: 26 September 2008
| MGD data statistics | 26 September 2008 |
|---|---|
| Genes with nucleotide sequence data | 28 869 |
| Genes with protein sequence data | 27 244 |
| Genes (including uncloned mutations) | 37 696 |
| Genes with gene traps | 12 390 |
| Mapped genes and markers | 46 288 |
| Genes with GO annotations | 18 082 |
| Mouse/human orthologs | 16 685 |
| Mouse/rat orthologs | 15 787 |
| Phenotypic alleles | 20 478 |
| Genes with one or more phenotypic alleles | 7876 |
| Phenotypic alleles that are targeted mutations | 12 338 |
| Genes with targeted mutations | 5306 |
| Human diseases with one or more mouse models | 858 |
| QTLs | 3979 |
| References | 133 867 |
| Mouse RefSNPs | 10 089 692 |
| Mouse nucleotide sequences integrated into the MGI system (includes ESTs) | >8 750 000 |
Only genes with nucleotide sequence data are included in the unified gene catalog.
2008 IMPROVEMENTS AND UPDATES
New ways to explore mouse phenotypes
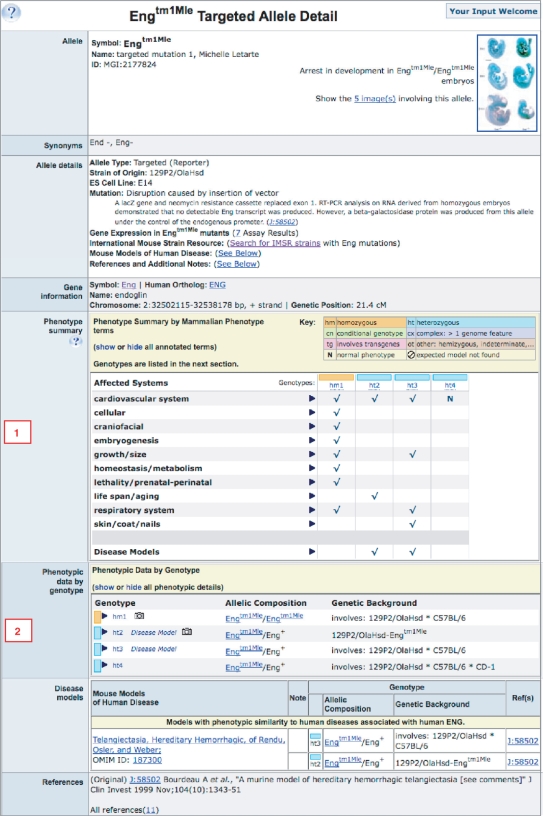
The Allele Detail page for each mutant allele in MGI now includes two distinct views of phenotype data that provide powerful options for exploring relationships between genotypes and phenotypes (Figure 1).
Figure 1.
Allele detail page for the Engtm1Mle targeted mutation. The ‘Phenotype summary’ section [labeled 1] displays a matrix view of phenotype terms (vertical axis) by genotypes (horizontal axis). Phenotype terms can be expanded to show more detail and each genotype abbreviation links to a page detailing the full phenotype for that genotype. The ‘Phenotypic data by genotype’ section [labeled 2] shows a table of genotypes involving Engtm1Mle. Each genotype can be expanded to reveal full phenotypes. All data for each of the phenotype sections of this page can be viewed using the ‘show’/‘hide’ options in the section headers.
In the ‘Phenotype summary’ section of the page, a matrix view of phenotypes (vertical axis) by genotypes (horizontal axis) allows users to quickly view the range of phenotypic effects observed for a given allele. The effects of different allelic combinations (such as homozygous, heterozygous, conditional and complex) in different genetic backgrounds can be compared. The general phenotype classes can be expanded individually (as shown in Figure 2A) or all phenotype terms can be viewed or hidden using the ‘show’/‘hide’ option in the matrix header. This matrix view can also be used to go directly to the phenotypic details for a specific genotype (displayed in a new window) by clicking on its genotype abbreviation (e.g. hm1, for homozygous 1).
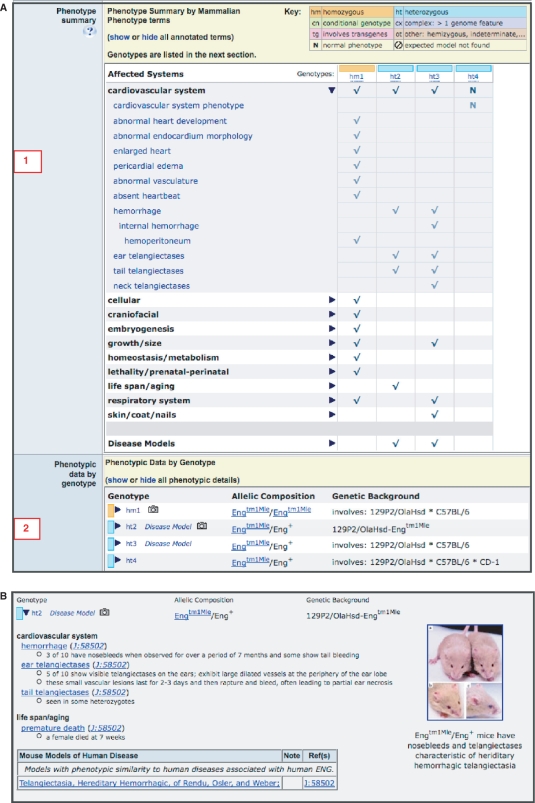
Figure 2.
Using the new expansion features for comparing phenotypes. (A) The ‘Phenotype summary’ matrix is shown expanded for the cardiovascular system term [labeled 1]. Note the finer granularity of the terms. The genetic background effect in Engtm1Mle/+heterozygotes can clearly be seen. Heterozygote 4 (ht4) displays a normal cardiovascular phenotype, compared with the other two heterozygous genotypes (ht2, ht3). By glancing below to the ‘Phenotypic data by genotype’ section [labeled 2], it can be observed that in Engtm1Mle/+mice, the addition of background alleles from the CD-1 strain appears to confer a protective effect for these cardiovascular system phenotypes. (B) The ‘Phenotypic data by genotype’ section is shown expanded for one of the genotypes (Engtm1Mle/+heterozygotes in the 129P2/OlaHsd-Engtm1Mle strain, abbreviated ht2).
The ‘Phenotypic data by genotype’ section presents a table of all genotypes involving the allele being viewed. Each genotype is a link that expands to reveal the full phenotype details for that genotype, including disease model associations (Figure 2B). Details for all genotypes containing the mutant allele can be viewed at once or hidden using the ‘show’/‘hide’ option in the header of this section.
A brief Allele Tour (http://www.informatics.jax.org/faq/Allele_tour.shtml) is available giving an overview of these changes and a help document further explains the Phenotypic Allele Detail pages (http://www.informatics.jax.org/userdocs/allele_detail_report.shtml).
Unified mouse gene catalog
The catalog of mouse genes in MGD serves as the foundation for functional annotation of all genes and genome features in the MGI database. The MGD gene curation process integrates gene predictions from Ensembl, NCBI and Vega into a single, nonredundant catalog. The unified gene catalog for most recent genome assembly (NCBI Build 37, or B37) is available from MGD and is updated when new gene predictions are released.
The concept of gene in the unified mouse gene catalog refers to the computational prediction of structural genome features including protein- and nonprotein-coding genes. The concept of gene in MGD generally includes the additional concept of heritable phenotype. That is, cases where an observable trait appears to be inherited in a typical Mendelian fashion but the underlying structural gene is not known.
Build 37 (B37), which includes ∼2.6 GB of mouse sequence, is considered to be ‘essentially complete’. MGD has the most current B37 data available from three providers, NCBI, Ensembl and Vega. The MGI Mouse Genome Sequencing group analyzed the files from these three sources to produce a unified mouse gene catalog that established associations between MGI markers and the updated coordinates. This allows researchers to obtain a comprehensive list of mouse genes from a single source and serves as the basis for functional annotation of genes in the MGI database.
The algorithm for our gene ‘unification’ process has been described previously (9). Rather than relying on sequence similarity to determine the equivalency of predicted genes, our process looks for the genome coordinate overlap of annotated exons. Combining the gene predictions from NCBI, Ensembl and Vega for B37 we produced a catalog of over 34 000 genes and pseudogenes in the mouse genome. Although the overlap of genes predicted by the different groups was significant there are also a large number of genes and pseudogenes that are unique to each of the gene prediction processes. For example, the initial analysis of gene predictions from B37 indicated that 6953 genes were unique to NCBI, 4707 were unique to Ensembl and 2986 were unique to Vega.
New web design and search tool
New web design
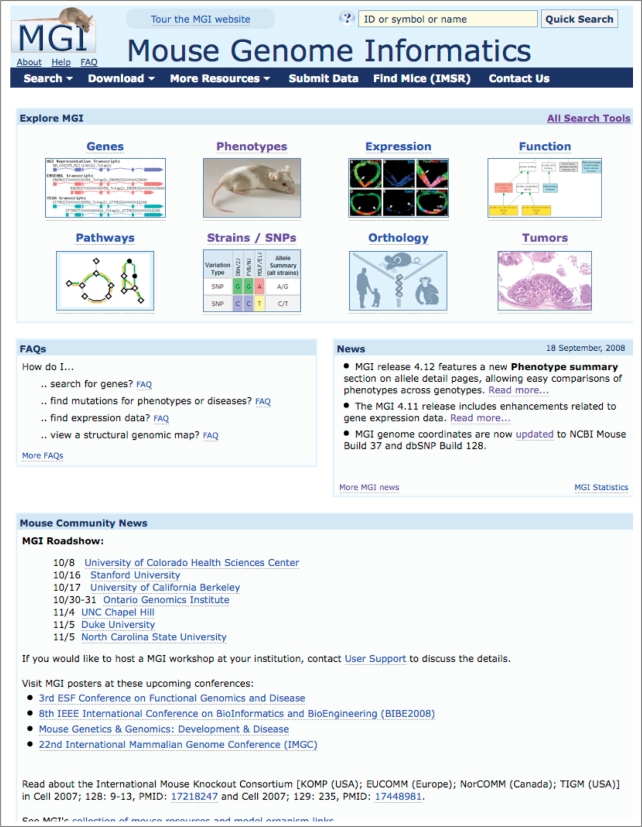
Exploring MGI is now assisted with a navigation bar that appears on each web page. The navigation bar features cascading menus that lead users quickly to specific search forms and information pages. The homepage (Figure 3) boasts new major content area images, leading to specific content pages that, in turn, provide relevant data access points and FAQs. This new navigation paradigm improves intuitive navigation of MGI, providing more visual clues for users and allowing quick access to the desired MGI pages.
Figure 3.
Redesigned MGI Homepage. Notable design items of the new MGI web pages include a navigation bar that is included on every MGI page, featuring cascading menus that lead users quickly to the query form or information page of interest. On the homepage, clickable images representing major content areas lead users to pages with additional information, descriptions of MGD data for that area, links to query forms and reports, and relevant FAQs.
New search tool
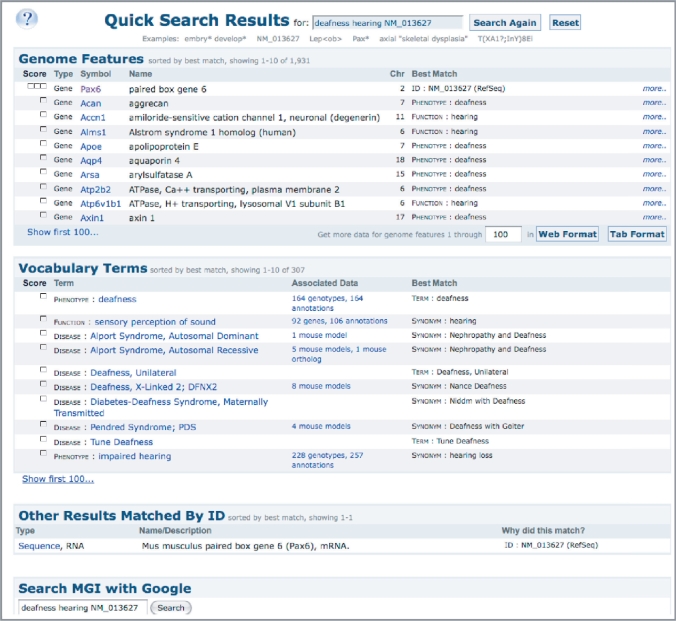
Recently, major infrastructure enhancements have made the MGI Quick Search Tool (Figure 4) a verbose and comprehensive search entrée into MGI data. The Quick Search now combines nomenclature and ID searches with searches of MGI annotations and ontologies. The combination of an enhanced nomenclature search (symbols, names, orthologs), and complete indexing of MGI data, and weighted word searches provides an instantaneous return of information, as well as data for the user on the nature of the returned object. The Quick Search has become a robust way for those unfamiliar with MGI to focus their interests and a simplified search for users who seek quick entry into specific information (e.g. give me detail for gene X; what information does MGI have about retinal degeneration?). Advanced search forms in MGI continue to support complex queries such as ‘What genes on Chromosome 11 functions as transcription factors and have mutations associated with abnormalities of the inner ear?’
Figure 4.
New MGI search tool. The new search tool provides maximum flexibility for quickly locating genes and annotations of interest in MGI. Searches are automatically done against nomenclature (gene symbols/names, synonyms, orthologs), ontologies/vocabularies used for MGI data associations, including gene function, process and cellular location (GO), phenotype (MP) and disease terms (OMIM), anatomical terms, protein domains (PIRSF) and accession IDs. Results returned are ranked by best match to the term(s) entered by the user and links are provided to the underlying data and to a comprehensive list of matches in the database. The figure shows the results for searching for: deafness hearing NM_013627. The terms deafness and/or hearing were matched to 267 genes and 246 vocabulary terms, and the sequence ID retrieved the corresponding RefSeq match.
COMMUNITY AUTHORITIES AND ACCESS
Mouse gene, allele and strain nomenclature
MGD is responsible for assigning official nomenclature to mouse genes, alleles and strains following the guidelines set by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/nomen). MGD staff work with various bioinformatics resource curators to resolve nomenclature inconsistencies resulting from regular data exchange of shared links, and with specialists for human (http://www.genenames.org/), rat (http://rgd.mcw.edu) and other species (e.g. zebrafish http://zfin.org) to provide an organized approach to the nomenclature process. Collaborative efforts between the mouse and human nomenclature committees and scientific experts in specific domain areas provide an up-to-date analysis and compilation of the latest knowledge about genes and gene families, such as the NLR family (10). The MGD group that also assists journal editors to ensure standardized nomenclature is adhered to in publications. The MGD nomenclature coordinator can be contacted by email (nomen@informatics.jax.org).
Electronic data submission
MGD accepts contributed data sets for any type of data maintained by the database. The most frequent types of contributed data are mutant allele and phenotypic information originating with the large mouse mutagenesis centers and repositories that contribute to the International Mouse Strain Resource (IMSR, http://www.imsr.org). Each electronic submission receives a permanent database accession ID. All data sets are associated with their source, either a publication or an electronic submission reference. Online details about data submission procedures is found at http://www.informatics.jax.org/mgihome/submissions/submissions_menu.shtml.
Community outreach and user support
MGD user support can be accessed through online documentation and easy email or phone access to User Support Staff.
World Wide Web:http://www.informatics.jax.org/mgihome/support/support.shtml
Email access:mgi-help@informatics.jax.org
Telephone access:1-207-288-6445
FAX access:1-207-288-6132
Other outreach: MGI-LIST (http://www.informatics.jax.org/mgihome/lists/lists.shtml) is a moderated and active email bulletin board supported by the MGD User Support group.
HIGH-LEVEL OVERVIEW OF THE MAIN COMPONENTS AND IMPLEMENTATION
MGD is implemented in the Sybase relational database management system with approximately 180 tables within which the biological information is stored. BLAST-able databases, genome assembly files for sequence data and image data are stored outside the relational database. An editing interface (EI) and automated load programs are used to input data into the MGD system. The EI is an interactive, graphical application used by curators. Automated load programs that integrate larger data sets from many sources into the database include quality control (QC) checks and processing algorithms that integrate the bulk of the data automatically and identify issues to be resolved by curators or the data provider. Thus, through EI and automated loads, we acquire and integrate large amounts of data into a high-quality, knowledgebase.
Public data access is provided through the web interface (WI) where users can interactively query and download our data through a web browser. MouseBLAST allows users to do sequence similarity searches against a variety of rodent-relevant sequence databases that are built weekly from selected sequence databases from NCBI, UniProt and other providers. Mouse GBrowse allows users to visualize mouse data sets against the genome as a series of linear tracks. FTP reports are a major source for other data providers who link to or use MGD data in their products, and for computational biologists who use MGD data in their analyses. Programmatic access to MGD via web services is also available. All MGD files and programs are openly and freely available.
CITING MGD
For a general citation of the MGD resource please cite this article. In addition, the following citation format is suggested when referring to datasets specific to the MGD component of MGI: Mouse Genome Database (MGD), Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine (URL: http://www.informatics.jax.org). [Type in date (month, year) when you retrieved the data cited.] Citation, Copyright, Warranty Disclaimer and other resource-specific information can be found in the footer of all MGI web pages.
FUNDING
NIH/NHGRI (grant HG000330 to Mouse Genome Database). Funding for open access charge: HG 000330.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bult CJ, Blake JA, Kadin JA, Richardson JE, Eppig JT the Mouse Genome Database Group. The Mouse Genome Database (MGD): mouse biology. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE the Mouse Genome Informatics Group. The Mouse Genome Database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake JA, Eppig JT, Bult CJ, Kadin JA, Richardson JE the Mouse Genome Database Group. The Mouse Genome Database (MGD): updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The Mouse Gene Expression Database (GXD): 2007 update. Nucleic Acids Res. 2007;35:D618–D623. doi: 10.1093/nar/gkl1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krupke DM, Begley DA, Sundberg JP, Bult CJ, Eppig JT. The Mouse Tumor Biology Database. Nat. Rev. Cancer. 2008;8:459–465. doi: 10.1038/nrc2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Gene Ontology Consortium. The Gene Ontology (GO) project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith CL, Goldsmith CA, Eppig JT. The mammalian phenotype ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson JE. Fjoin: simple and efficient computation of feature overlaps. J. Comput. Biol. 2006;13:1457–1464. doi: 10.1089/cmb.2006.13.1457. [DOI] [PubMed] [Google Scholar]
- 10.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]