Abstract
MMsINC (http://mms.dsfarm.unipd.it/MMsINC/search) is a database of non-redundant, richly annotated and biomedically relevant chemical structures. A primary goal of MMsINC is to guarantee the highest quality and the uniqueness of each entry. MMsINC then adds value to these entries by including the analysis of crucial chemical properties, such as ionization and tautomerization processes, and the in silico prediction of 24 important molecular properties in the biochemical profile of each structure. MMsINC is consequently a natural input for different chemoinformatics and virtual screening applications. In addition, MMsINC supports various types of queries, including substructure queries and the novel ‘molecular scissoring’ query. MMsINC is interfaced with other primary data collectors, such as PubChem, Protein Data Bank (PDB), the Food and Drug Administration database of approved drugs and ZINC.
INTRODUCTION
One of the most important issues in the post-proteomic era is relating protein pharmacology by ligand chemistry (1). In fact, there is intriguing pharmacological evidence that related drugs can recognize molecular targets that appear unrelated by many bioinformatics metrics (1). Drug side effects and related toxicity profiles can be considered the obvious consequence of this polypharmacology. The capability of chemically related drugs to bind proteins without sequence or structure similarity can limit attempts in bioinformatics to understand and categorize their pharmacological action. On the other hand, a chemo-centric approach to this problem compares not only the biological targets themselves, but also the chemistry involving their ligands (1,2).
We have planned MMsINC with this chemo-centric approach in mind, integrating chemical structures, their chemical behaviour annotations and purpose-specific search functions, with the aim of creating a valuable tool for the interpretation of protein pharmacology (including toxicology) by ligand chemistry. (If you refer to MMsINC database and web interface for your published research, we ask that you please cite this article.) Like others, from a chemoinformatics point of view MMsINC is a chemical structure database, where chemicals are appropriately stored and annotated. However, MMsINC has as its main priorities to eliminate redundancy in its data, and to guarantee the accuracy of all chemical annotations derived from a chemical structure to avoid chemical misleading. An accurate chemical annotation is also crucial for the significance of any qualitative or quantitative chemical similarity metrics, which are a key principle in ligand/drug design and a good guide to the biological comparison of chemicals (3). Indeed, structure- and property-based similarities are useful tools implemented by MMsINC with the goal to establish chemical connections among all MMsINC entries and other publicly available databases (PDB (4), PubChem (5), DrugBank (6), ZINC (7) and ChemDB (8), among others). We also apply MMsINC's structure-based annotation and similarity to classify all MMsINC entries into sets of several biologically relevant targets based on their fragment-driven similarity score. MMsINC is consequently a natural input for different chemoinformatics and virtual screening applications, and its integration with well-consolidated virtual screening tools, such as pharmacophore screening and molecular docking is in progress (Table 1).
Table 1.
Comparison of MMsINC with other publicly available molecular databases (Y = available)
| ZINC | ChemBank | PubChem | ChemDB | MMsINC | |
|---|---|---|---|---|---|
| Features | |||||
| Full download | Y | Ya | Y | Y | |
| Subset download | Y | Ya | Y | Y | Y |
| Size | 8M | 1.7M | 19.6M | 5M | 4M |
| Non-redundant (InChI-based) | Y | ||||
| Chirality | Y | Y | Y | Y | |
| FDA similarities | Y | ||||
| PDB ligand similarities | Y | ||||
| Link to PDB | Y | Y | Y | ||
| Link to PubChem | N/A | Y | Y | ||
| Search | |||||
| By exact structure | Y | Y | Y | ||
| By similarity | Y | Y | Y | Y | |
| By substructure | Y | Y | Y | Y | Y |
| By fragment | Y | ||||
| By molecular descriptors | Y | Y | Y | Y | Y |
| By assays | Y | Y |
aOnly for registered users.
All of MMsINC's data and functions are accessible through a user-friendly web interface that we describe later in this text.
DATABASE CREATION
MMsINC is a public, web-based informatics platform derived from the aggregation and multi-step treatment of 46 data sources: primarily commercial vendor catalogues, but also of publicly available repositories (e.g. NCI, http://cactus.nci.nih.gov). For sources that periodically update their data and make them available on the Internet, we automatically download the data and synchronize MMsINC with the latest version. Complete information about all the vendors is available in the Supplementary Materials. In total, the current database contains about 4 million unique compounds, resulting from the distillation of the original set of 7.5 million entries (Table 2).
Table 2.
MMsINC Database statistics. Drug- and lead-likeness have been measured by Lipinski (11) and Oprea (12) criteria
| Frameworks | more than 175,000 |
|---|---|
| Drug-like molecules | 3.89 million (98%) |
| Lead-like molecules | 3.61 million (91%) |
| Chemically stable compounds | 3.45 million (87%) |
The objective of our treatment process is to generate a data set that is free of redundancy and with a chemical orthography that is as accurate as possible. In addition, we calculate the most probable tautomeric and ionic states at physiological conditions, and we produce one possible stable conformer for each molecular entry. These are the steps we follow to assemble the MMsINC data set.
Step 1: first redundancy washing
Using the Molecular Operating Environment software suite (MOE, version 2007.09, http://www.chemcomp.com), we remove all originally redundant entries based on their SMILES (9) representation, reducing the number of entries from 7.5 M to 4 M.
Step 2: generation of tautomers
We apply the LigPrep 2.1 tool by Schrödinger LLC (http://www.schrodinger.com/) to generate the tautomers for each molecule resulting from Step 1. Many of the tautomers we generate at this stage are unstable molecules, and therefore unlikely to be encountered in practice. These will be eliminated later in our process. In the meantime, we add all the generated tautomers to our data set.
Step 3: generation of ionic states
By ionic states, we refer to the most energetically favourable electrically charged states that a molecule can assume at a pH of 7.4. We calculate the most favourable ionic states for each molecular entry resulting from Step 2 by using the ‘Protonate’ tool in the MOE suite. The calculations add a further 250 000 MMsINC entries.
Step 4: conformer selection
The three-dimensional (3D) structure of each MMsINC entry (including all tautomers and ionic states) is calculated by using Corina 3.4 (http://www.mol-net.de). For each input structure, Corina generates possible conformers, which are variations of the molecule where the existing bonds are the same, but parts of the molecule are rotated differently along these bonds. The software then selects one of the lowest energy conformers generated. We use Corina with its default configuration.
Step 5: second redundancy washing
The SMILES encoding used for the first redundancy washing is ambiguous, making it possible for some molecular redundancy to slip through Step 1. In addition, it is possible for Steps 2 and 3 to generate identical structures. We therefore perform a second, InChI-based (10), redundancy washing that eliminates any such duplicates and achieves our goal of structural uniqueness in the database.
Step 6: unstable tautomer elimination
To eliminate the unstable tautomers generated in Step 2, we determine their energy stability with a force field-based criterion using the 3D structural data calculated in Step 4. For each ‘parent’ neutral molecule, we then keep in our data set only the most stable tautomers, up to a maximum of five. Instead, the less stable tautomers are discarded. The calculations leave us with ∼1.1 M tautomers that become part of the MMsINC data set.
The result of these steps is a non-redundant data set of neutral molecular structures, as well as their stable tautomers and ionic states.
Molecular descriptors
For the molecules resulting from the data set building process, we calculate 24 molecular properties useful for quantitative structure–activity relationship (QSAR), diversity analysis or combinatorial library design. We assign the corresponding partial charges to all atoms of each unique conformer by using the MMFF94 force field algorithm implemented by MOE. All other descriptors summarized in Table 3 are calculated using the MOE tool ‘QSAR-Descriptor’. For a detailed explanation of the descriptors, please refer to the MMsINC help pages accessible via Internet.
Table 3.
MMsINC molecular descriptors
| Descriptor Category | Descriptors |
|---|---|
| Physical | Molecular weight, reactive groups, SlogP, logS |
| Topological | Globularity, Sterimol/B1-4/L |
| Surface and Volume | ASA/+/−/H, Volume |
| Pharmacophoric | HB donor, HB acceptor, acid and basic groups, chiral centers |
| Energetic | Potential energy |
| Drug candidacy | Lipinski drug-like, Oprea lead-like |
QUERYING THE DATABASE
The MMsINC database is accessible to the public via our web application. It allows users to search the database by structural criteria, either by specifying a structure by one of the standard notations (SMILES, InChI, standard molecular formula), by drawing it with the Java Molecular Editor (JME, by Peter Ertl, http://www.molinspiration.com/jme/) or by identifying a structure in the MMsINC database by its MMsCode—our database's unique molecular identifier. The application also allows users to find MMsINC molecules by similarity to PDB Ligands (4), which in turn are selected by similarity to a query structure. In the following sections, we describe these different structure and similarity search methods supported by MMsINC.
Identical structure search
The identical structure search allows the user to search for molecules that match the structure specified by the query. MMsCode and InChI queries will result in at most one result, since they are unambiguous. On the other hand, SMILES and molecular formulas are ambiguous, so the search will return all molecules represented by the query found in MMsINC.
Substructure search
The substructure search is a query that allows the user to find molecules that contain a substructure of interest. The user specifies the substructure as a SMILES string or as an MMsCode which identifies a structure known to MMsINC.
The query is executed using structural keys, which are bit vectors that indicate with a ‘1’ the presence of a particular structural feature, and with a ‘0’ its absence. MMsINC uses 643-bit structural keys that identify a subset of the structures in the PubChem (5) fingerprints. If the query is specified as a SMILES string, MMsINC generates a query structural key dynamically. On the other hand, if the query is an MMsCode the system fetches the precalculated structural key associated with the identified molecule. The query key is then compared with the structural keys of the molecules in MMsINC, and the molecules that contain all the structural bits of the query are retrieved. This method provides a rapid and effective screening of the database, but its results can include false positives, since the key may not completely describe the query structure. Therefore, these preliminary results are filtered by an exact subgraph containment check using the Chemistry Development Kit (CDK) library (13).
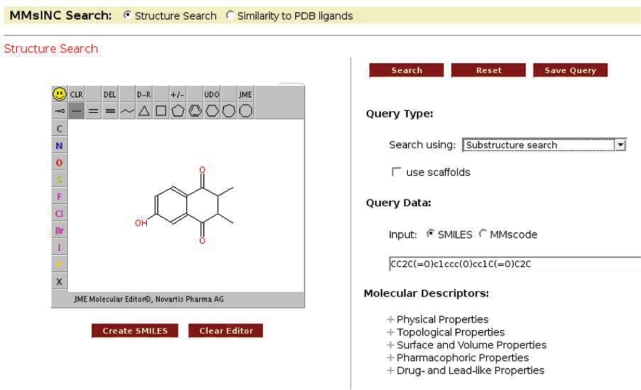
In some cases the preliminary key search results in too many molecules to perform the subgraph containment check on all the molecules in a reasonable amount of time. In these cases, MMsINC only performs the subgraph isomorphism check on the molecules as they are displayed to the user, indicating whether they are false positives. Currently this behaviour is applied only if the structural key search results in more than 30 000 results (Figure 1).
Figure 1.
MMsINC's substructure search screen.
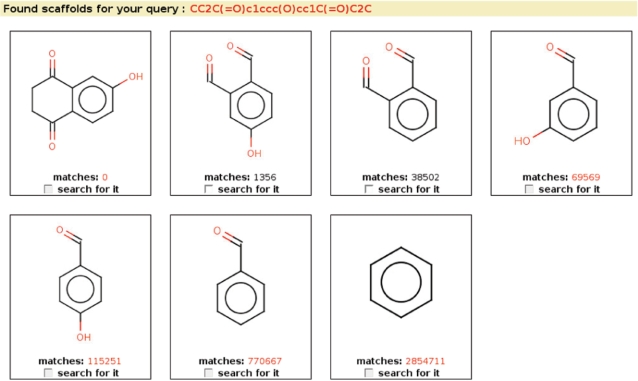
Molecular scissoring search
The molecular scissoring search is a novel type of query based on chemically relevant molecular fragments known as scaffolds. The user provides a query structure, as in the substructure search described in the previous section. The system identifies the scaffolds present in the query structure, and asks the user to specify which ones to use for the search, and whether to perform an ‘and’ query searching for the molecules that contain all the selected scaffolds, or an ‘or’ query searching for the molecules that contain any of the selected scaffolds. The query is then sent to the database where it is evaluated with the help of appropriate indices over the scaffold data.
Our current implementation of the scissoring search can in some rare cases allow the user to select scaffolds that do not exist in the query molecule. We know about this issue and are already working to resolve it. However, we do not believe it is serious enough to justify disabling the scissoring query in MMsINC. This query type is important because it allows chemists to easily and quickly search for molecules that contain particular chemical substructures that are known to be chemically and/or pharmacologically relevant (Figure 2).
Figure 2.
Scaffold selection screen.
Similarity search
The similarity search is a query type that retrieves all molecules in MMsINC that are structurally similar to query molecule. We measure the similarity between structures using the Tanimoto similarity score (3) on the structural keys describing them. The Tanimoto similarity is defined as the ratio of the number of bits set to ‘1’ in both keys to the number of bits set to ‘1’ in either of the two keys. For two structural keys A, B we have:
To perform a similarity search, the user enters a query structure and specifies a minimum acceptable similarity score. MMsINC compares the precalculated structural keys of its molecules with the structural key of the query and returns all the molecules with a Tanimoto similarity greater than or equal to the threshold.
To accelerate Tanimoto similarity searches, we have implemented the technique by Swamidass and Baldi (14). Their result allows us to bound the number of ones in the target structural key required to achieve a similarity score that meets the threshold, considerably reducing the number of molecules for which we actually need to calculate the Tanimoto similarity. The fraction of the database that is pruned from the search by using this technique varies from 30% to 100%.
Filtering by descriptors
As a supplement to the more sophisticated structural query methods just described, MMsINC users can further filter their search results (except when performing the identical structure search) by the molecular descriptors that MMsINC provides for each molecule in the database.
Displaying structural query results
Results from a structural query are displayed in pages of up to 20 molecules. For each result, the system displays the structural diagram of the molecule and its MMsCode. Users can select results that interest them and place them in the MMsINC ‘cart’ as they browse through the search results. The cart can then be saved locally to a standard SDF file.
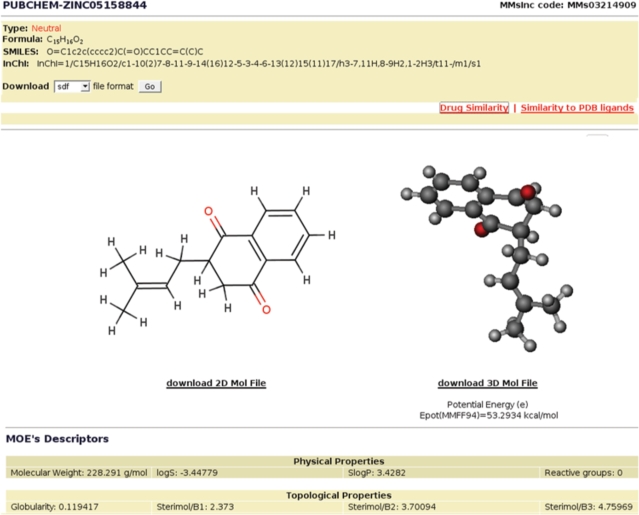
Clicking on a molecule from the results list takes the user to the molecule report for the specific selection (Figure 3). The report shows the user basic information about the molecule like the compound type (neutral, tautomer or ionic state), the molecular formula and its InChI and SMILES representations. The report also contains a 2D image of the molecule, and a 3D-movable rendering of molecule shown using Chemis3D (http://chemis.free.fr/mol3d/) Java applet. In addition, the precalculated descriptors for the molecule are listed at the bottom of the report. Finally, for neutral molecules the system lists all its tautomers and ions, while for tautomers and ions the neutral state of the molecule is indicated.
Figure 3.
Report for molecule MMs03214909.
From the molecule's report page the user can download the structural and descriptor data to his or her own computer in several standardized formats, including SDF, PDB, XYZ, as well as 2D and 3D MOLFILE. The user can also retrieve the list of the PDB ligands that are similar to the molecule, as well as retrieving a list of structurally similar FDA-approved drugs. Finally, the report has links to the PubChem and ZINC entries for the molecule.
Query by similarity to PDB ligands
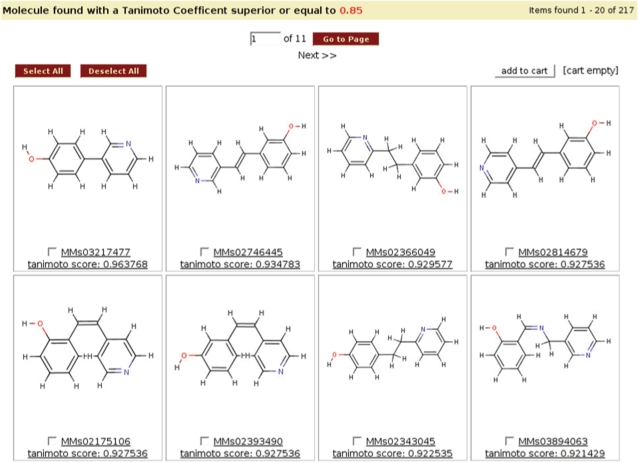
An alternative method to query MMsINC is via similarity to PDB ligands that have been inserted into our database. This search type has two modes of operation: SMILES and PDB. In the first mode, the user provides a molecular structure as a query, and a minimum threshold for the Tanimoto similarity measure. MMsINC finds PDB ligands with a Tanimoto similarity to the query structure greater than or equal to the specified threshold. In the second mode, the user specifies a list of up to five PDB protein identifiers. MMsINC finds all the ligands for the identified PDB proteins. In either case, MMsINC presents to the user the identified PDB ligands, with their structural diagram and the ligand code. Selecting a specific ligand takes the user to the ligand report page.
The ligand report page summarizes all the MMsINC neutral molecules, tautomers, ionic states and FDA-approved drugs that are similar to the ligand, with a Tanimoto similarity score threshold specified by the user but ≥0.70 (Figure 4). Clicking on any of these molecules takes the user to the molecule's report page. The ligand report page also contains basic information about the ligand, such as its 2D structural diagram, its three-letter code and its name, and a table showing all the PDB proteins that interact with this ligand is displayed with a click.
Figure 4.
The results page from an MMsINC query.
The PDB ligand search is based on the complete set of ligands from the PDB Chemical Component Dictionary (CCD). The MMsINC database integrates the version of the PDB CCD retrieved on January 31, 2008. However, it should be noted that not all of these PDB ligands used by the query mechanism are included in the main MMsINC data.
Implementation
The MMsINC system uses the PostgreSQL RDBMS (http://www.postgresql.org) to manage its data. The database is installed on a server running Linux.
The system's web application has been developed in PHP, with some components written in Java. MMsINC uses the CDK to perform some of its molecular analyses.
CONCLUSION
The MMsINC database has been created to support a chemo-centric approach to relate protein pharmacology by ligand chemistry. The primary aims of this growing database are the accuracy of all chemical information and the elimination of redundant data. In addition, MMsINC is naturally predisposed to integrate chemical, biological and pharmacological information coming from other publicly available databases. Finally, its useful and novel molecule query functions make it a new tool for chemoinformaticians. The integration with consolidated virtual screening tools, such as pharmacophore screening and molecular docking, will be available in the next release of MMsINC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Sardegna Ricerche through the project ‘Progetto Cluster Biomedicina e Tecnologie della Salute, sottoprogetto Bioinformatica’; the CRS4 (Bioinformatics Program); the Italian Ministry for University and Research (MIUR); University of Padova, Italy. Funding for open access charge: Sardegna Ricerche, Regional Government Agency for research and innovation.
Conflict of interest statement: none declared.
ACKNOWLEDGEMENTS
Stefano Moro is very grateful to the Chemical Computing Group, Schrödinger and Molecular Networks for the long and fruitful collaborations.
REFERENCES
- 1.Kaiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber SL. Small molecules: the missing link in the central dogma. Nat. Chem. Biol. 2005;1:64–66. doi: 10.1038/nchembio0705-64. [DOI] [PubMed] [Google Scholar]
- 3.Johnson MA, Maggiora GM. Concepts and Applications of Molecular Similarity. New York: Wiley; 1990. [Google Scholar]
- 4.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JH, Linstead E, Swamidass SJ, Wang D, Baldi P. ChemDB update–full-text search and virtual chemical space. Bioinformatics. 2007;23:2348–2351. doi: 10.1093/bioinformatics/btm341. [DOI] [PubMed] [Google Scholar]
- 9.Anderson E, Veith GD, Weininger D. Report No. EPA/600/M-87/021. Duluth, MN: U.S. EPA, Environmental Research Laboratory-Duluth; 1987. SMILES: a line notation and computerized interpreter for chemical structures. [Google Scholar]
- 10.McNaught A. The IUPAC International Chemical Identifier: InChI. Chem. Int. 2006;28:12–15. doi: 10.1186/s13321-015-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 12.Oprea TI. Property distribution of drug-related chemical databases. J. Comp. Aid. Mol. Des. 2000;14:251–264. doi: 10.1023/a:1008130001697. [DOI] [PubMed] [Google Scholar]
- 13.Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Recent developments of the Chemistry Development Kit (CDK)—an open-source Java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 14.Swamidass SJ, Baldi P. Bounds and algorithms for fast exact searches of chemical fingerprints in linear and sublinear time. J. Chem. Inf. Model. 2007;47:302–317. doi: 10.1021/ci600358f. [DOI] [PMC free article] [PubMed] [Google Scholar]